2RBE
 
 | | The discovery of 2-anilinothiazolones as 11beta-HSD1 inhibitors | | Descriptor: | (5R)-2-[(2-fluorophenyl)amino]-5-(1-methylethyl)-1,3-thiazol-4(5H)-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, J, Jordan, S.R, Li, V. | | Deposit date: | 2007-09-18 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The discovery of 2-anilinothiazolones as 11beta-HSD1 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6IUR
 
 | | A phosphatase complex STRN3-PP2Aa | | Descriptor: | PP2A scaffolding subunit, PROPANE, Striatin-3 | | Authors: | Tang, Y, Zhou, Z.C. | | Deposit date: | 2018-11-30 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Selective Inhibition of STRN3-Containing PP2A Phosphatase Restores Hippo Tumor-Suppressor Activity in Gastric Cancer.
Cancer Cell, 38, 2020
|
|
8JAY
 
 | | CrtSPARTA Octamer bound with guide-target | | Descriptor: | DNA (25-MER), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Guo, L.J, Huang, P.P, Li, Z.X, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-05-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
4LY9
 
 | | Human GKRP complexed to AMG-1694 [(2R)-1,1,1-trifluoro-2-{4-[(2S)-2-{[(3S)-3-methylmorpholin-4-yl]methyl}-4-(thiophen-2-ylsulfonyl)piperazin-1-yl]phenyl}propan-2-ol] and sorbitol-6-phosphate | | Descriptor: | (2R)-1,1,1-trifluoro-2-{4-[(2S)-2-{[(3S)-3-methylmorpholin-4-yl]methyl}-4-(thiophen-2-ylsulfonyl)piperazin-1-yl]phenyl}propan-2-ol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Jordan, S.R. | | Deposit date: | 2013-07-30 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Antidiabetic effects of glucokinase regulatory protein small-molecule disruptors.
Nature, 504, 2013
|
|
7TBF
 
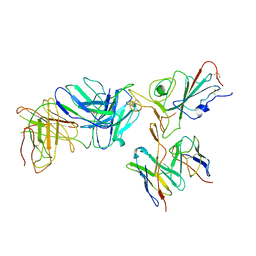 | | Locally refined region of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of SARS-CoV-2 antibody A19-61.1, Heavy chain of SARS-CoV-2 antibody B1-182.1, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.
Science, 376, 2022
|
|
7TB8
 
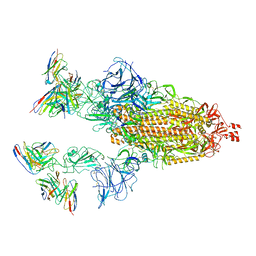 | | Cryo-EM structure of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.
Science, 376, 2022
|
|
7TCC
 
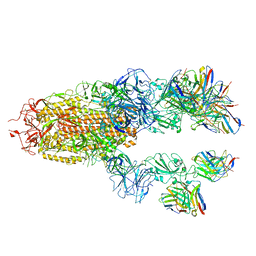 | |
7TC9
 
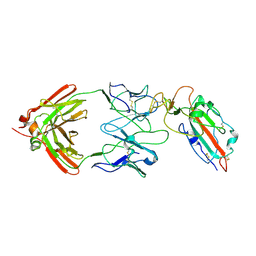 | |
7TCA
 
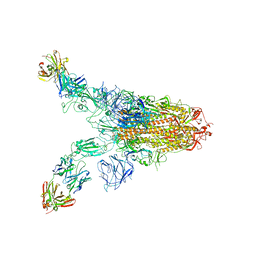 | | Cryo-EM structure of SARS-CoV-2 Omicron spike in complex with antibody A19-46.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of antibody A19-46.1, ... | | Authors: | Zhou, T, Kwong, P.D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.
Science, 376, 2022
|
|
7U0D
 
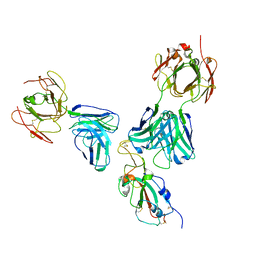 | |
4MMS
 
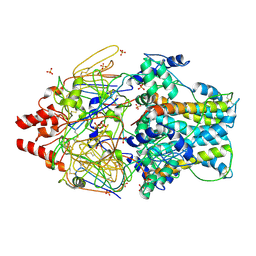 | | Crystal Structure of Prefusion-stabilized RSV F Variant Cav1 at pH 5.5 | | Descriptor: | Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2, SULFATE ION | | Authors: | Mclellan, J.S, Joyce, M.G, Stewart-Jones, G.B.E, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2013-09-09 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
4MMR
 
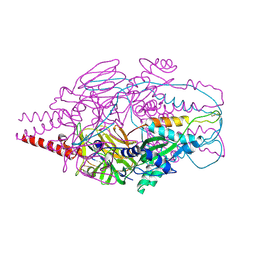 | | Crystal Structure of Prefusion-stabilized RSV F Variant Cav1 at pH 9.5 | | Descriptor: | Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2 | | Authors: | Stewart-Jones, G.B.E, McLellan, J.S, Joyce, M.G, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2013-09-09 | | Release date: | 2013-11-20 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
4MMV
 
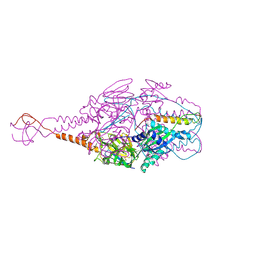 | | Crystal Structure of Prefusion-stabilized RSV F Variant DS-Cav1-TriC at pH 9.5 | | Descriptor: | Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2 | | Authors: | Stewart-Jones, G.B.E, McLellan, J.S, Joyce, M.G, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2013-09-09 | | Release date: | 2013-11-20 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
4MMU
 
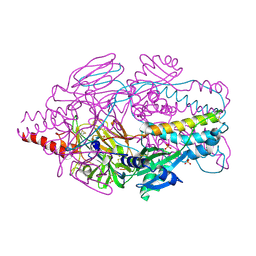 | | Crystal Structure of Prefusion-stabilized RSV F Variant DS-Cav1 at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2, ... | | Authors: | Joyce, M.G, Mclellan, J.S, Stewart-Jones, G.B.E, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2013-09-09 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
4MMT
 
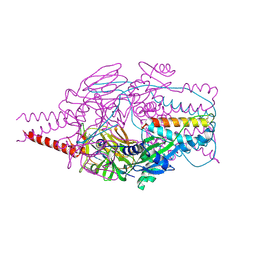 | | Crystal Structure of Prefusion-stabilized RSV F Variant DS-Cav1 at pH 9.5 | | Descriptor: | Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2 | | Authors: | Joyce, M.G, Mclellan, J.S, Stewart-Jones, G.B.E, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2013-09-09 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
4MMQ
 
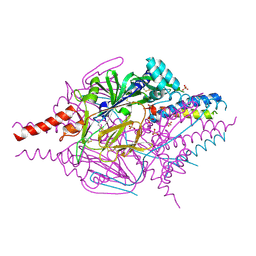 | | Crystal Structure of Prefusion-stabilized RSV F Variant DS | | Descriptor: | Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2, SULFATE ION | | Authors: | Mclellan, J.S, Joyce, M.G, Stewart-Jones, G.B.E, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2013-09-09 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.253 Å) | | Cite: | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
3BYZ
 
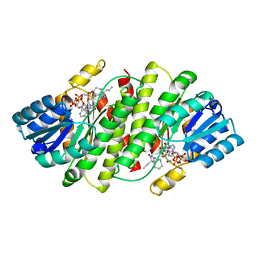 | | 2-Amino-1,3-thiazol-4(5H)-ones as Potent and Selective 11-Hydroxysteroid Dehydrogenase Type 1 Inhibitors | | Descriptor: | (5S)-2-(cyclooctylamino)-5-methyl-5-propyl-1,3-thiazol-4(5H)-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, J, Jordan, S.R, Li, V. | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | 2-amino-1,3-thiazol-4(5H)-ones as potent and selective 11beta-hydroxysteroid dehydrogenase type 1 inhibitors: enzyme-ligand co-crystal structure and demonstration of pharmacodynamic effects in C57Bl/6 mice.
J.Med.Chem., 51, 2008
|
|
3CHQ
 
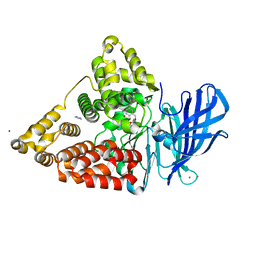 | | Crystal structure of leukotriene a4 hydrolase in complex with N5-[4-(phenylmethoxy)phenyl]-L-glutamine | | Descriptor: | (2S)-2-amino-5-oxo-5-[(4-phenylmethoxyphenyl)amino]pentanoic acid, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
3CHR
 
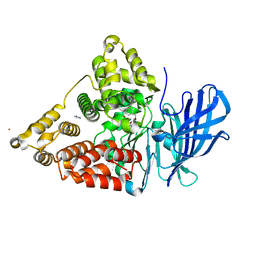 | | Crystal structure of leukotriene A4 hydrolase in complex with 4-amino-N-[4-(phenylmethoxy)phenyl]-butanamide | | Descriptor: | 4-amino-N-[4-(benzyloxy)phenyl]butanamide, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
4N6J
 
 | | Crystal structure of human Striatin-3 coiled coil domain | | Descriptor: | Striatin-3 | | Authors: | Chen, C, Shi, Z, Zhou, Z. | | Deposit date: | 2013-10-13 | | Release date: | 2014-02-26 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Striatins contain a noncanonical coiled coil that binds protein phosphatase 2A A subunit to form a 2:2 heterotetrameric core of striatin-interacting phosphatase and kinase (STRIPAK) complex.
J.Biol.Chem., 289, 2014
|
|
8WD8
 
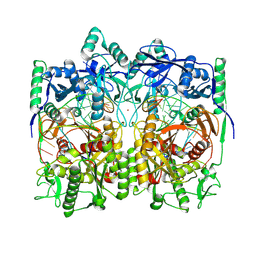 | | Cryo-EM structure of TtdAgo-guide DNA-target DNA complex | | Descriptor: | Argonaute family protein, Guide DNA, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute.
Mol.Cell, 84, 2024
|
|
3CHS
 
 | | Crystal structure of leukotriene A4 hydrolase in complex with (2S)-2-amino-5-[[4-[(2S)-2-hydroxy-2-phenyl-ethoxy]phenyl]amino]-5-oxo-pentanoic acid | | Descriptor: | (2S)-2-amino-5-[[4-[(2S)-2-hydroxy-2-phenyl-ethoxy]phenyl]amino]-5-oxo-pentanoic acid, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
3CHO
 
 | | Crystal structure of leukotriene a4 hydrolase in complex with 2-amino-N-[4-(phenylmethoxy)phenyl]-acetamide | | Descriptor: | ACETATE ION, Leukotriene A-4 hydrolase, N-[4-(benzyloxy)phenyl]glycinamide, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
3CHP
 
 | | Crystal structure of leukotriene a4 hydrolase in complex with (3S)-3-amino-4-oxo-4-[(4-phenylmethoxyphenyl)amino]butanoic acid | | Descriptor: | (3S)-3-amino-4-oxo-4-[(4-phenylmethoxyphenyl)amino]butanoic acid, ACETATE ION, IMIDAZOLE, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
5W23
 
 | | Crystal Structure of RSV F in complex with 5C4 Fab | | Descriptor: | 5C4 Fab heavy chain, 5C4 Fab light chain, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S. | | Deposit date: | 2017-06-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of respiratory syncytial virus subtype-dependent neutralization by an antibody targeting the fusion glycoprotein.
Nat Commun, 8, 2017
|
|
