5CSB
 
 | | The crystal structure of beta2-microglobulin D76N mutant at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
5CS7
 
 | | The crystal structure of wt beta2-microglobulin at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
3IB4
 
 | |
3JQP
 
 | |
3JQR
 
 | |
1QHA
 
 | | HUMAN HEXOKINASE TYPE I COMPLEXED WITH ATP ANALOGUE AMP-PNP | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Rosano, C, Sabini, E, Deriu, D, Magnani, M, Bolognesi, M. | | Deposit date: | 1999-05-11 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Binding of non-catalytic ATP to human hexokinase I highlights the structural components for enzyme-membrane association control.
Structure Fold.Des., 7, 1999
|
|
3JQQ
 
 | |
2QRW
 
 | | Crystal structure of Mycobacterium tuberculosis trHbO WG8F mutant | | Descriptor: | CYANIDE ION, Hemoglobin-like protein HbO, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Milani, M, Bolognesi, M. | | Deposit date: | 2007-07-30 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Roles of Tyr(CD1) and Trp(G8) in Mycobacterium tuberculosis Truncated Hemoglobin O in Ligand Binding and on the Heme Distal Site Architecture
Biochemistry, 46, 2007
|
|
2Z9T
 
 | | Crystal structure of the human beta-2 microglobulin mutant W60G | | Descriptor: | Beta-2-microglobulin | | Authors: | Ricagno, S, Bolognesi, M, Bellotti, V, Corazza, A, Rennella, E, Gural, D, Mimmi, M.C, Betto, E, Pucillo, C, Fogolari, F, Viglino, P, Raimondi, S, Giorgetti, S, Bolognesi, B, Merlini, G, Stoppini, M. | | Deposit date: | 2007-09-26 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The controlling roles of Trp60 and Trp95 in beta2-microglobulin function, folding and amyloid aggregation properties
J.Mol.Biol., 378, 2008
|
|
7ZCF
 
 | |
7ZCE
 
 | |
2OXT
 
 | | Crystal structure of Meaban virus nucleoside-2'-O-methyltransferase | | Descriptor: | NUCLEOSIDE-2'-O-METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | Authors: | Mastrangelo, E, Milani, M, Bollati, M, Bolognesi, M. | | Deposit date: | 2007-02-21 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural bases for substrate recognition and activity in Meaban virus nucleoside-2'-O-methyltransferase
Protein Sci., 16, 2007
|
|
5N2C
 
 | | Crystal structure of the peptidoglycan-associated lipoprotein from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ACETATE ION, ... | | Authors: | Matterazzo, E, Bolognesi, M, Gourlay, L.J. | | Deposit date: | 2017-02-07 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designing Probes for Immunodiagnostics: Structural Insights into an Epitope Targeting Burkholderia Infections.
ACS Infect Dis, 3, 2017
|
|
8A11
 
 | | Cryo-EM structure of the Human SHMT1-RNA complex | | Descriptor: | SHMT2 5'UTR 1-50 (5'-R(P*CP*UP*AP*CP*AP*A)-3'), Serine hydroxymethyltransferase, cytosolic | | Authors: | Spizzichino, S, Marabelli, C, Bharadwaj, A, Jakobi, A.J, Chaves-Sanjuan, A, Giardina, G, Bolognesi, M, Cutruzzola, F. | | Deposit date: | 2022-05-30 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Cryo-EM structure of the Human SHMT1-RNA complex
To Be Published
|
|
3QZX
 
 | | 3D Structure of ferric methanosarcina acetivorans protoglobin Y61A mutant with unknown ligand | | Descriptor: | GLYCEROL, Methanosarcina acetivorans protoglobin, PHOSPHATE ION, ... | | Authors: | Pesce, A, Tilleman, L, Dewilde, S, Ascenzi, P, Coletta, M, Ciaccio, C, Bruno, S, Moens, L, Bolognesi, M, Nardini, M. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural heterogeneity and ligand gating in ferric methanosarcina acetivorans protoglobin mutants.
Iubmb Life, 63, 2011
|
|
1ZN2
 
 | | Low Resolution Structure of Response Regulator StyR | | Descriptor: | MAGNESIUM ION, response regulatory protein | | Authors: | Milani, M, Leoni, L, Rampioni, G, Zennaro, E, Ascenzi, P, Bolognesi, M. | | Deposit date: | 2005-05-11 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | An Active-like Structure in the Unphosphorylated StyR Response Regulator Suggests a Phosphorylation- Dependent Allosteric Activation Mechanism.
STRUCTURE, 13, 2005
|
|
2C0K
 
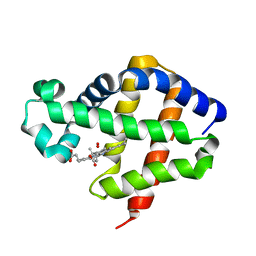 | | The structure of hemoglobin from the botfly Gasterophilus intestinalis | | Descriptor: | HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Hoogewijs, D, Ascenzi, P, Moens, L, Bolognesi, M. | | Deposit date: | 2005-09-05 | | Release date: | 2005-11-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Modulation of Oxygen Binding to Insect Hemoglobins: The Structure of Hemoglobin from the Botfly Gasterophilus Intestinalis.
Protein Sci., 14, 2005
|
|
2QEQ
 
 | |
4RMW
 
 | |
4RMU
 
 | |
4RMV
 
 | |
2IG3
 
 | | Crystal structure of group III truncated hemoglobin from Campylobacter jejuni | | Descriptor: | ACETATE ION, CYANIDE ION, Group III truncated haemoglobin, ... | | Authors: | Nardini, M, Pesce, A, Labarre, M, Ascenzi, P, Guertin, M, Bolognesi, M. | | Deposit date: | 2006-09-22 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural determinants in the group III truncated hemoglobin from Campylobacter jejuni.
J.Biol.Chem., 281, 2006
|
|
3OZ1
 
 | | cIAP1-BIR3 domain in complex with the Smac-mimetic compound Smac066 | | Descriptor: | (3S,6S,7R,9aS)-7-[2-(benzylamino)ethyl]-N-(diphenylmethyl)-6-{[(2S)-2-(methylamino)butanoyl]amino}-5-oxooctahydro-1H-pyrrolo[1,2-a]azepine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Cossu, F, Malvezzi, F, Mastrangelo, E, Canevari, G, Bolognesi, M, Milani, M. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recognition of Smac-mimetic compounds by the BIR3 domain of cIAP1
Protein Sci., 2010
|
|
1SDY
 
 | | STRUCTURE SOLUTION AND MOLECULAR DYNAMICS REFINEMENT OF THE YEAST CU,ZN ENZYME SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Djinovic, K, Gatti, G, Coda, A, Antolini, L, Pelosi, G, Desideri, A, Falconi, M, Marmocchi, F, Rotilio, G, Bolognesi, M. | | Deposit date: | 1991-06-14 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure solution and molecular dynamics refinement of the yeast Cu,Zn enzyme superoxide dismutase.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
3QJ4
 
 | | Crystal structure of Human Renalase (isoform 1) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Renalase, SULFATE ION | | Authors: | Milani, M, Ciriello, F, Baroni, S, Pandini, V, Aliverti, A, Canevari, G, Bolognesi, M. | | Deposit date: | 2011-01-28 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | FAD-binding site and NADP reactivity in human renalase: a new enzyme involved in blood pressure regulation
J.Mol.Biol., 411, 2011
|
|
