2HHQ
 
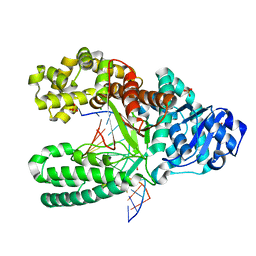 | | O6-methyl-guanine:T pair in the polymerase-10 basepair position | | Descriptor: | 5'-D(*CP*AP*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*AP*TP*GP*A)-3', DNA polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HHU
 
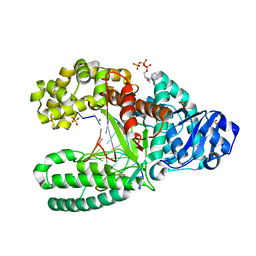 | | C:O6-methyl-guanine in the polymerase postinsertion site (-1 basepair position) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*GP*CP*GP*AP*TP*CP*AP*GP*CP*CP*C)-3', 5'-D(*GP*TP*AP*CP*(6OG)P*GP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3', ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HHV
 
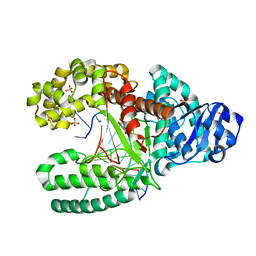 | | T:O6-methyl-guanine in the polymerase-2 basepair position | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*AP*GP*CP*TP*TP*G)-3', 5'-D(*GP*TP*AP*CP*(6OG)P*AP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3', DNA Polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HHX
 
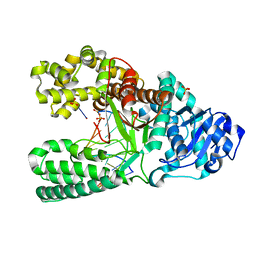 | | O6-methyl-guanine in the polymerase template preinsertion site | | Descriptor: | 5'-D(*CP*AP*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*G)-3', DNA Polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HVH
 
 | | ddCTP:O6MeG pair in the polymerase active site (0 position) | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*CP*A*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DDG))-3', ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-07-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HVI
 
 | | ddCTP:G pair in the polymerase active site (0 position) | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*C*AP*TP*GP*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DDG))-3', ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-07-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HW3
 
 | | T:O6-methyl-guanine pair in the polymerase postinsertion site (-1 basepair position) | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*AP*GP*CP*TP*T)-3', 5'-D(*GP*TP*A*CP*(6OG)P*AP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3', DNA Polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-07-31 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2IEJ
 
 | | Human Protein Farnesyltransferase Complexed with Inhibitor Compound STN-48 And FPP Analog at 1.8A Resolution | | Descriptor: | ACETATE ION, METHYL N-{(3S)-1-[(1-METHYL-1H-IMIDAZOL-5-YL)METHYL]-6-PHENYL-1,2,3,4-TETRAHYDROQUINOLIN-3-YL}-N-[(1-METHYL-1H-IMIDAZOL-4-YL)SULFONYL]GLYCINATE, Protein farnesyltransferase subunit beta, ... | | Authors: | Hast, M.A, Beese, L.S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-01-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Resistance mutations at the lipid substrate binding site of Plasmodium falciparum protein farnesyltransferase.
Mol.Biochem.Parasitol., 152, 2007
|
|
1QBQ
 
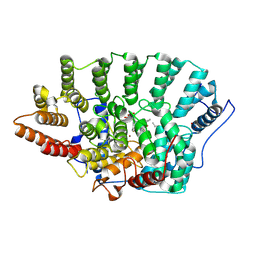 | | STRUCTURE OF RAT FARNESYL PROTEIN TRANSFERASE COMPLEXED WITH A CVIM PEPTIDE AND ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID. | | Descriptor: | ACETATE ION, ACETYL-CYS-VAL-ILE-SELENOMET-COOH PEPTIDE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, ... | | Authors: | Strickland, C.L, Windsor, W.T, Syto, R, Wang, L, Bond, R, Wu, Z, Schwartz, J, Le, H.V, Beese, L.S, Weber, P.C. | | Deposit date: | 1999-04-27 | | Release date: | 1999-06-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of farnesyl protein transferase complexed with a CaaX peptide and farnesyl diphosphate analogue
Biochemistry, 37, 1998
|
|
1MZC
 
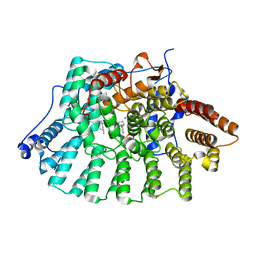 | | Co-Crystal Structure Of Human Farnesyltransferase With Farnesyldiphosphate and Inhibitor Compound 33a | | Descriptor: | 2-[3-(3-ETHYL-1-METHYL-2-OXO-AZEPAN-3-YL)-PHENOXY]-4-[1-AMINO-1-(1-METHYL-1H-IMIDIZOL-5-YL)-ETHYL]-BENZONITRILE, FARNESYL DIPHOSPHATE, Protein Farnesyltransferase alpha Subunit, ... | | Authors: | deSolms, S.J, Ciccarone, T.M, MacTough, S.C, Shaw, A.W, Buser, C.A, Ellis-Hutchings, M, Fernandes, C, Hamilton, K.A, Huber, H.E, Kohl, N.E, Lobell, R.B, Robinson, R.G, Tsou, N.N, Walsh, E.S, Graham, S.L, Beese, L.S, Taylor, J.S. | | Deposit date: | 2002-10-07 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual Protein Farnesyltransferase-Geranylgeranyltransferase-I Inhibitors as Potential Cancer Chemotherapeutic Agents.
J.Med.Chem., 46, 2003
|
|
1N4R
 
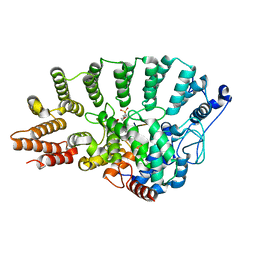 | | Protein Geranylgeranyltransferase type-I Complexed with a Geranylgeranylated KKKSKTKCVIL Peptide Product | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Fusion protein consisting of transforming protein p21b and Ras related protein Rap-2b, ... | | Authors: | Taylor, J.S, Reid, T.S, Casey, P.J, Beese, L.S. | | Deposit date: | 2002-11-01 | | Release date: | 2003-11-18 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of mammalian protein geranylgeranyltransferase type-I
EMBO J., 22, 2003
|
|
1N4P
 
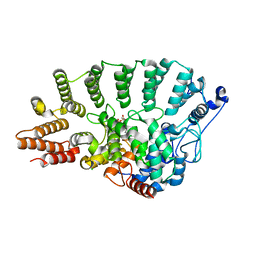 | | Protein Geranylgeranyltransferase type-I Complexed with Geranylgeranyl Diphosphate | | Descriptor: | CHLORIDE ION, Fusion protein consisting of transforming protein p21b and Ras related protein Rap-2b, GERAN-8-YL GERAN, ... | | Authors: | Taylor, J.S, Reid, T.S, Casey, P.J, Beese, L.S. | | Deposit date: | 2002-11-01 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of mammalian protein geranylgeranyltransferase type-I
EMBO J., 22, 2003
|
|
1N4Q
 
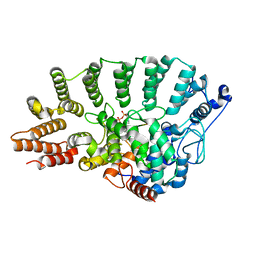 | | Protein Geranylgeranyltransferase type-I Complexed with a GGPP Analog and a KKKSKTKCVIL Peptide | | Descriptor: | 2-[METHYL-(5-GERANYL-4-METHYL-PENT-3-ENYL)-AMINO]-ETHYL-DIPHOSPHATE, CHLORIDE ION, Fusion protein consisting of transforming protein p21b and Ras related protein Rap-2b, ... | | Authors: | Taylor, J.S, Reid, T.S, Casey, P.J, Beese, L.S. | | Deposit date: | 2002-11-01 | | Release date: | 2003-11-18 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of mammalian protein geranylgeranyltransferase type-I
EMBO J., 22, 2003
|
|
1N4S
 
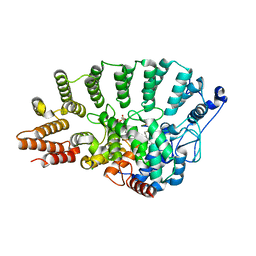 | | Protein Geranylgeranyltransferase type-I Complexed with GGPP and a Geranylgeranylated KKKSKTKCVIL Peptide Product | | Descriptor: | CHLORIDE ION, Fusion protein consisting of transforming protein p21b and Ras related protein Rap-2b, GERAN-8-YL GERAN, ... | | Authors: | Taylor, J.S, Reid, T.S, Casey, P.J, Beese, L.S. | | Deposit date: | 2002-11-01 | | Release date: | 2003-11-18 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of mammalian protein geranylgeranyltransferase type-I
EMBO J., 22, 2003
|
|
1S63
 
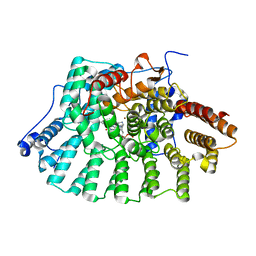 | | Human protein farnesyltransferase complexed with L-778,123 and FPP | | Descriptor: | 4-[(5-{[4-(3-CHLOROPHENYL)-3-OXOPIPERAZIN-1-YL]METHYL}-1H-IMIDAZOL-1-YL)METHYL]BENZONITRILE, FARNESYL DIPHOSPHATE, Protein farnesyltransferase beta subunit, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Analysis Reveals that Anticancer Clinical Candidate L-778,123 Inhibits Protein Farnesyltransferase and Geranylgeranyltransferase-I by Different Binding Modes.
Biochemistry, 43, 2004
|
|
1SA5
 
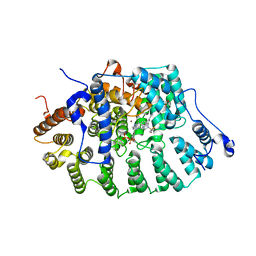 | | Rat protein farnesyltransferase complexed with FPP and BMS-214662 | | Descriptor: | 3-BENZYL-1-(1H-IMIDAZOL-4-YLMETHYL)-4-(THIEN-2-YLSULFONYL)-2,3,4,5-TETRAHYDRO-1H-1,4-BENZODIAZEPINE-7-CARBONITRILE, ACETIC ACID, FARNESYL DIPHOSPHATE, ... | | Authors: | Reid, T.S, Beese, L.S. | | Deposit date: | 2004-02-06 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Anticancer Clinical Candidates R115777 (Tipifarnib) and BMS-214662 Complexed with Protein Farnesyltransferase Suggest a Mechanism of FTI Selectivity.
Biochemistry, 43, 2004
|
|
1S64
 
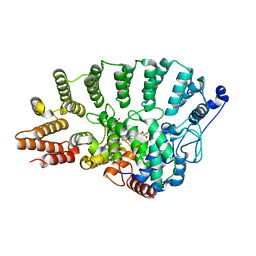 | | Rat protein geranylgeranyltransferase type-I complexed with L-778,123 and a sulfate anion | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[(5-{[4-(3-CHLOROPHENYL)-3-OXOPIPERAZIN-1-YL]METHYL}-1H-IMIDAZOL-1-YL)METHYL]BENZONITRILE, CHLORIDE ION, ... | | Authors: | Reid, T.S, Long, S.B, Beese, L.S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystallographic Analysis Reveals that Anticancer Clinical Candidate L-778,123 Inhibits Protein Farnesyltransferase and Geranylgeranyltransferase-I by Different Binding Modes.
Biochemistry, 43, 2004
|
|
1SA4
 
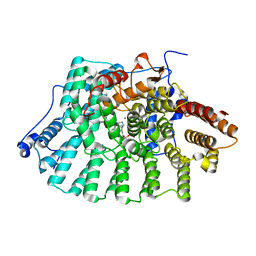 | | human protein farnesyltransferase complexed with FPP and R115777 | | Descriptor: | 6-[(S)-AMINO(4-CHLOROPHENYL)(1-METHYL-1H-IMIDAZOL-5-YL)METHYL]-4-(3-CHLOROPHENYL)-1-METHYLQUINOLIN-2(1H)-ONE, FARNESYL DIPHOSPHATE, Protein farnesyltransferase beta subunit, ... | | Authors: | Reid, T.S, Beese, L.S. | | Deposit date: | 2004-02-06 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Anticancer Clinical Candidates R115777 (Tipifarnib) and BMS-214662 Complexed with Protein Farnesyltransferase Suggest a Mechanism of FTI Selectivity.
Biochemistry, 43, 2004
|
|
4YFU
 
 | |
3I5O
 
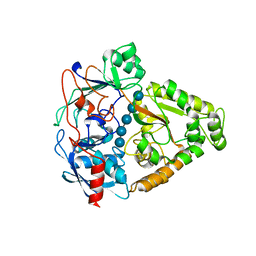 | | The X-ray crystal structure of a thermophilic cellobiose binding protein bound with cellopentaose | | Descriptor: | Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cuneo, M.J, Hellinga, H.W. | | Deposit date: | 2009-07-06 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of Semi-specific Oligosaccharide Recognition by a Cellulose-binding Protein of Thermotoga maritima Reveals Adaptations for Functional Diversification of the Oligopeptide Periplasmic Binding Protein Fold.
J.Biol.Chem., 284, 2009
|
|
2O7I
 
 | | The X-ray crystal structure of a thermophilic cellobiose binding protein bound with cellobiose | | Descriptor: | Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cuneo, M.J, Hellinga, H.W. | | Deposit date: | 2006-12-11 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of Semi-specific Oligosaccharide Recognition by a Cellulose-binding Protein of Thermotoga maritima Reveals Adaptations for Functional Diversification of the Oligopeptide Periplasmic Binding Protein Fold.
J.Biol.Chem., 284, 2009
|
|
2FN8
 
 | |
2FNC
 
 | | Thermotoga maritima maltotriose binding protein bound with maltotriose. | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose ABC transporter, periplasmic maltose-binding protein | | Authors: | Cuneo, M.J, Changela, A. | | Deposit date: | 2006-01-10 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The
crystal structure of redundant maltotriose binding proteins from the
thermophile Thermotoga maritima
To be Published
|
|
2H3H
 
 | |
2GX6
 
 | |
