1XYA
 
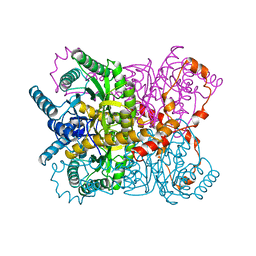 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | Descriptor: | HYDROXIDE ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-01-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
8DB9
 
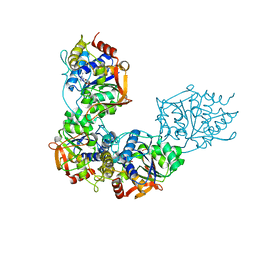 | | Adenosine/guanosine nucleoside hydrolase bound to inhibitor | | Descriptor: | 1-beta-D-ribofuranosyl-1H-1,2,4-triazole-3-carboximidamide, CALCIUM ION, Inosine-uridine preferring nucleoside hydrolase family protein | | Authors: | Muellers, S.N, Allen, K.N, Stockman, B.J. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-Guided Insight into the Specificity and Mechanism of a Parasitic Nucleoside Hydrolase.
Biochemistry, 61, 2022
|
|
8DB6
 
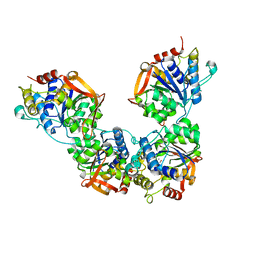 | | Adenosine/guanosine nucleoside hydrolase | | Descriptor: | CALCIUM ION, GLYCEROL, Inosine-uridine preferring nucleoside hydrolase family protein | | Authors: | Muellers, S.N, Allen, K.N, Stockman, B.J. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-Guided Insight into the Specificity and Mechanism of a Parasitic Nucleoside Hydrolase.
Biochemistry, 61, 2022
|
|
8DB8
 
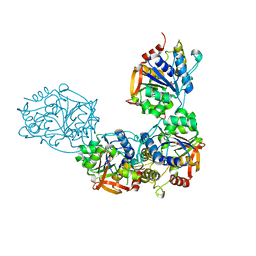 | | Adenosine/guanosine nucleoside hydrolase bound to ImH | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, Inosine-uridine preferring nucleoside hydrolase family protein | | Authors: | Muellers, S.N, Allen, K.N, Stockman, B.J. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure-Guided Insight into the Specificity and Mechanism of a Parasitic Nucleoside Hydrolase.
Biochemistry, 61, 2022
|
|
8DB7
 
 | | Adenosine/guanosine nucleoside hydrolase bound to a fragment inhibitor | | Descriptor: | CALCIUM ION, GLYCEROL, Inosine-uridine preferring nucleoside hydrolase family protein, ... | | Authors: | Muellers, S.N, Allen, K.N, Stockman, B.J. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure-Guided Insight into the Specificity and Mechanism of a Parasitic Nucleoside Hydrolase.
Biochemistry, 61, 2022
|
|
7N18
 
 | | Clostridium botulinum Neurotoxin Serotype A Light Chain Inhibited by a Chiral Hydroxamic Acid | | Descriptor: | (3R)-3-(4-chlorophenyl)-N,5-dihydroxypentanamide, (3S)-3-(4-chlorophenyl)-N,5-dihydroxypentanamide, Botulinum neurotoxin type A, ... | | Authors: | Silvaggi, N.R, Allen, K.N. | | Deposit date: | 2021-05-27 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Use of Crystallography and Molecular Modeling for the Inhibition of the Botulinum Neurotoxin A Protease.
Acs Med.Chem.Lett., 12, 2021
|
|
1CQD
 
 | | THE 2.1 ANGSTROM STRUCTURE OF A CYSTEINE PROTEASE WITH PROLINE SPECIFICITY FROM GINGER RHIZOME, ZINGIBER OFFICINALE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (PROTEASE II), ... | | Authors: | Choi, K.H, Laursen, R.A, Allen, K.N. | | Deposit date: | 1999-06-15 | | Release date: | 1999-09-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1 A structure of a cysteine protease with proline specificity from ginger rhizome, Zingiber officinale.
Biochemistry, 38, 1999
|
|
1U7P
 
 | | X-ray Crystal Structure of the Hypothetical Phosphotyrosine Phosphatase MDP-1 of the Haloacid Dehalogenase Superfamily | | Descriptor: | MAGNESIUM ION, TUNGSTATE(VI)ION, magnesium-dependent phosphatase-1 | | Authors: | Peisach, E, Selengut, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Crystal Structure of the Hypothetical Phosphotyrosine
Phosphatase MDP-1 of the Haloacid Dehalogenase Superfamily
Biochemistry, 43, 2004
|
|
1U7O
 
 | | Magnesium Dependent Phosphatase 1 (MDP-1) | | Descriptor: | ACETATE ION, magnesium-dependent phosphatase-1 | | Authors: | Peisach, E, Selengut, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystal structure of the hypothetical phosphotyrosine phosphatase MDP-1 of the haloacid dehalogenase superfamily
Biochemistry, 43, 2004
|
|
5TTK
 
 | |
5TTJ
 
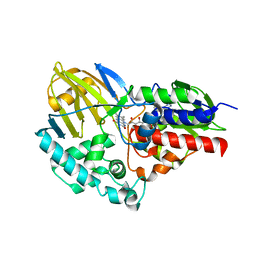 | |
3E81
 
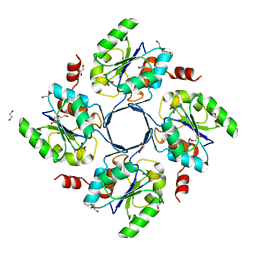 | | Structure-function Analysis of 2-Keto-3-deoxy-D-glycero-D-galacto-nononate-9-phosphate (KDN) Phosphatase Defines a New Clad Within the Type C0 HAD Subfamily | | Descriptor: | 1,2-ETHANEDIOL, Acylneuraminate cytidylyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lu, Z, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2008-08-19 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structure-Function Analysis of 2-Keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate Phosphatase Defines Specificity Elements in Type C0 Haloalkanoate Dehalogenase Family Members.
J.Biol.Chem., 284, 2009
|
|
3E8M
 
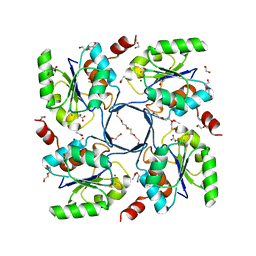 | | Structure-function Analysis of 2-Keto-3-deoxy-D-glycero-D-galacto-nononate-9-phosphate (KDN) Phosphatase Defines a New Clad Within the Type C0 HAD Subfamily | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Acylneuraminate cytidylyltransferase, ... | | Authors: | Lu, Z, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2008-08-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure-Function Analysis of 2-Keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate Phosphatase Defines Specificity Elements in Type C0 Haloalkanoate Dehalogenase Family Members.
J.Biol.Chem., 284, 2009
|
|
3LTQ
 
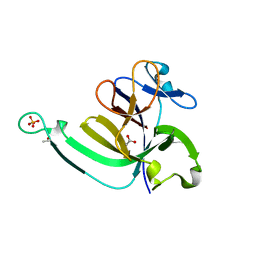 | |
3E84
 
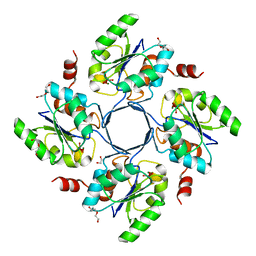 | | Structure-function Analysis of 2-Keto-3-deoxy-D-glycero-D-galacto-nononate-9-phosphate (KDN) Phosphatase Defines a New Clad Within the Type C0 HAD Subfamily | | Descriptor: | 1,2-ETHANEDIOL, Acylneuraminate cytidylyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lu, Z, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2008-08-19 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Function Analysis of 2-Keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate Phosphatase Defines Specificity Elements in Type C0 Haloalkanoate Dehalogenase Family Members.
J.Biol.Chem., 284, 2009
|
|
2OJR
 
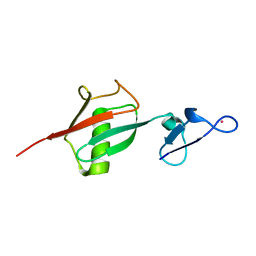 | |
5WFL
 
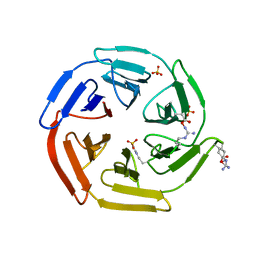 | | Kelch domain of human Keap1 in open unliganded conformation | | Descriptor: | Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Carolan, J.P, Lynch, A.J, Allen, K.N. | | Deposit date: | 2017-07-12 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Interaction Energetics and Druggability of the Protein-Protein Interaction between Kelch-like ECH-Associated Protein 1 (KEAP1) and Nuclear Factor Erythroid 2 Like 2 (Nrf2).
Biochemistry, 59, 2020
|
|
5WH9
 
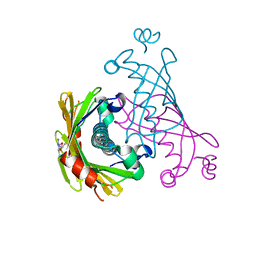 | |
5WHL
 
 | |
5WIY
 
 | | Kelch domain of human Keap1 bound to small molecule inhibitor fragment: 4-amino-1,7-dihydro-6H-pyrazolo[3,4-d]pyrimidine-6-thione | | Descriptor: | 4-amino-1,7-dihydro-6H-pyrazolo[3,4-d]pyrimidine-6-thione, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Carolan, J.P, Lynch, A.J, Allen, K.N. | | Deposit date: | 2017-07-20 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Interaction Energetics and Druggability of the Protein-Protein Interaction between Kelch-like ECH-Associated Protein 1 (KEAP1) and Nuclear Factor Erythroid 2 Like 2 (Nrf2).
Biochemistry, 59, 2020
|
|
5WG1
 
 | | Kelch domain of human Keap1 bound to mutant Nrf2 EAGE peptide | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 EAGE mutant peptide, SULFATE ION | | Authors: | Carolan, J.P, Lynch, A.J, Allen, K.N. | | Deposit date: | 2017-07-13 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.021 Å) | | Cite: | Interaction Energetics and Druggability of the Protein-Protein Interaction between Kelch-like ECH-Associated Protein 1 (KEAP1) and Nuclear Factor Erythroid 2 Like 2 (Nrf2).
Biochemistry, 59, 2020
|
|
5WFV
 
 | | Kelch domain of human Keap1 bound to Nrf2 ETGE peptide | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 ETGE peptide, SULFATE ION | | Authors: | Carolan, J.P, Lynch, A.J, Allen, K.N. | | Deposit date: | 2017-07-12 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Interaction Energetics and Druggability of the Protein-Protein Interaction between Kelch-like ECH-Associated Protein 1 (KEAP1) and Nuclear Factor Erythroid 2 Like 2 (Nrf2).
Biochemistry, 59, 2020
|
|
5WHO
 
 | |
6P2D
 
 | |
4JDP
 
 | | Crystal structure of probable p-nitrophenyl phosphatase (pho2) (target EFI-501307) from Archaeoglobus fulgidus DSM 4304 with Magnesium bound | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, p-nitrophenyl phosphatase (Pho2) | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Allen, K.N, Dunaway-Mariano, D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of probable p-nitrophenyl phosphatase from Archaeoglobus fulgidus.
To be Published
|
|
