8XAW
 
 | |
8XAY
 
 | |
8WB6
 
 | |
3KF4
 
 | |
3M3Z
 
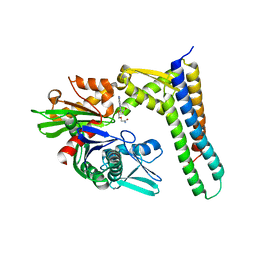 | | Crystal structure of HSC70/BAG1 in complex with small molecule inhibitor | | Descriptor: | 5'-O-(2-amino-2-oxoethyl)-8-(methylamino)adenosine, BAG family molecular chaperone regulator 1, Heat shock cognate 71 kDa protein | | Authors: | Dokurno, P, Surgenor, A.E, Shaw, T, Macias, A.T, Massey, A.J, Williamson, D.S. | | Deposit date: | 2010-03-10 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Adenosine-Derived Inhibitors of 78 kDa Glucose Regulated Protein (Grp78) ATPase: Insights into Isoform Selectivity.
J.Med.Chem., 54, 2011
|
|
7CBO
 
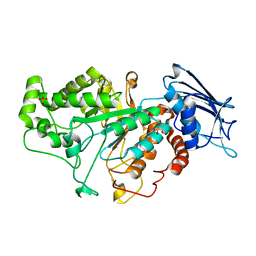 | | Crystal structure of beta-N-acetylhexosaminidase Am0868 from Akkermansia muciniphila in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, GLYCEROL, ... | | Authors: | Xu, W, Wang, M, Zhang, M. | | Deposit date: | 2020-06-13 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and biochemical analyses of beta-N-acetylhexosaminidase Am0868 from Akkermansia muciniphila involved in mucin degradation.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
5CQJ
 
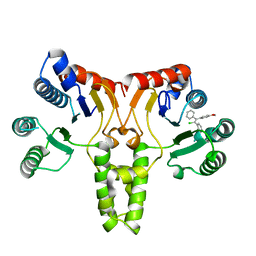 | | Crystal structure of E. coli undecaprenyl pyrophosphate synthase in complex with clomiphene | | Descriptor: | Clomifene, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) | | Authors: | Worrall, L.J, Conrady, D.G, Strynadka, N.C. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Antagonism screen for inhibitors of bacterial cell wall biogenesis uncovers an inhibitor of undecaprenyl diphosphate synthase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7CBN
 
 | |
5CQB
 
 | | Crystal structure of E. coli undecaprenyl pyrophosphate synthase | | Descriptor: | Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific), TRIETHYLENE GLYCOL | | Authors: | Worrall, L.J, Conrady, D.G, Strynadka, N.C. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antagonism screen for inhibitors of bacterial cell wall biogenesis uncovers an inhibitor of undecaprenyl diphosphate synthase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3MO8
 
 | | PWWP Domain of Human Bromodomain and PHD finger-containing protein 1 In Complex with Trimethylated H3K36 Peptide | | Descriptor: | Histone H3.2 TRIMETHYLATED H3K36 PEPTIDE, Peregrin | | Authors: | Lam, R, Zeng, H, Ni, S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
3M06
 
 | | Crystal Structure of TRAF2 | | Descriptor: | TNF receptor-associated factor 2 | | Authors: | Kabaleeswaran, V, Wu, H. | | Deposit date: | 2010-03-02 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structures of the TRAF2: cIAP2 and the TRAF1: TRAF2: cIAP2 complexes: affinity, specificity, and regulation.
Mol.Cell, 38, 2010
|
|
8WB7
 
 | |
3L42
 
 | | PWWP domain of human bromodomain and PHD finger containing protein 1 | | Descriptor: | Peregrin, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Zeng, H, Ni, S, Amaya, M.F, Dong, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
6X1C
 
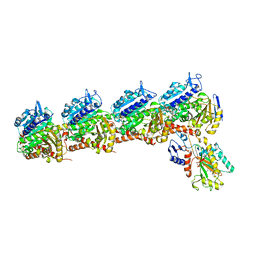 | | Tubulin-RB3_SLD-TTL in complex with compound 5j | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-chlorofuro[3,2-d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
6X1F
 
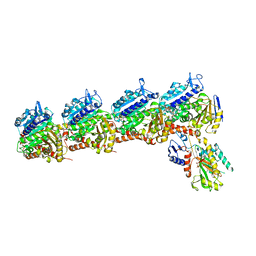 | | Tubulin-RB3_SLD-TTL in complex with compound 5m | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-methoxy-4-(2-methyl-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-yl)-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
6X1E
 
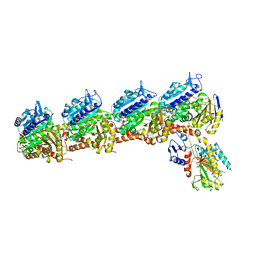 | | Tubulin-RB3_SLD-TTL in complex with compound 5l | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-chloro-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
7B54
 
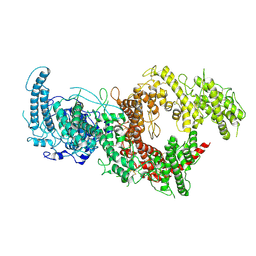 | | VAR2CSA full ectodomain in present of plCS, DBL1-DBL4 | | Descriptor: | VAR2CSA in presence of plCS, DBl1-DBL4,Erythrocyte membrane protein 1 | | Authors: | Wang, K.T, Dagil, R, Gourdon, P.E, Salanti, A. | | Deposit date: | 2020-12-03 | | Release date: | 2021-06-02 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM reveals the architecture of placental malaria VAR2CSA and provides molecular insight into chondroitin sulfate binding.
Nat Commun, 12, 2021
|
|
3MTF
 
 | | Crystal structure of the ACVR1 kinase in complex with a 2-aminopyridine inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-[6-amino-5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenol, Activin receptor type-1, ... | | Authors: | Chaikuad, A, Sanvitale, C, Cooper, C, Mahajan, P, Daga, N, Petrie, K, Alfano, I, Canning, P, Krojer, T, Vollmar, M, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A new class of small molecule inhibitor of BMP signaling.
Plos One, 8, 2013
|
|
7CLI
 
 | | Structure of NF-kB p52 homodimer bound to P-Selectin kB DNA fragment | | Descriptor: | DNA (5'-D(*CP*AP*AP*GP*GP*GP*GP*TP*CP*AP*CP*CP*CP*CP*CP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*GP*GP*GP*GP*GP*TP*GP*AP*CP*CP*CP*CP*TP*TP*G)-3'), Nuclear factor NF-kappa-B p52 subunit | | Authors: | Meshcheryakov, V.A, Wang, V.Y.-F. | | Deposit date: | 2020-07-21 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of NF-kappa B p52 homodimer-DNA complexes rationalize binding mechanisms and transcription activation.
Elife, 12, 2023
|
|
3NCO
 
 | | Crystal structure of FnCel5A from F. nodosum Rt17-B1 | | Descriptor: | Endoglucanase FnCel5A, PHOSPHATE ION, peptide (ALA)(ASN)(GLU), ... | | Authors: | Zheng, B.S, Yang, W, Wang, Y, Lou, Z.Y, Rao, Z.H, Feng, Y. | | Deposit date: | 2010-06-05 | | Release date: | 2011-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of FnCel5A from F. nodosum Rt17-B1
To be Published
|
|
6KWY
 
 | | human PA200-20S complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Proteasome activator complex subunit 4, Proteasome subunit alpha type-1, ... | | Authors: | Ouyang, S, Hongxin, G. | | Deposit date: | 2019-09-09 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures.
Plos Biol., 18, 2020
|
|
5XRO
 
 | |
5XRI
 
 | |
5XRJ
 
 | |
5XSU
 
 | | novel orally efficacious inhibitors complexed with PARP1 | | Descriptor: | 6-fluoranyl-2-(4,5,6,7-tetrahydrofuro[2,3-c]pyridin-2-yl)-1~{H}-benzimidazole-4-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Liu, Q, Xu, Y. | | Deposit date: | 2017-06-15 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of 2-(4,5,6,7-tetrahydrothienopyridin-2-yl)-benzoimidazole carboxamides as novel orally efficacious Poly(ADP-ribose)polymerase (PARP) inhibitors
Eur J Med Chem, 145, 2018
|
|
