6MG2
 
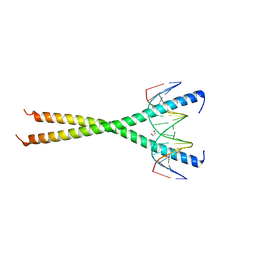 | | C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence-C2221 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.928 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
5DEM
 
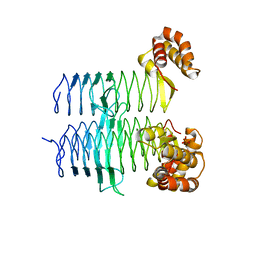 | | Structure of Pseudomonas aeruginosa LpxA | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, PHOSPHATE ION | | Authors: | Smith, E.W, Chen, Y. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structures of Pseudomonas aeruginosa LpxA Reveal the Basis for Its Substrate Selectivity.
Biochemistry, 54, 2015
|
|
6VC8
 
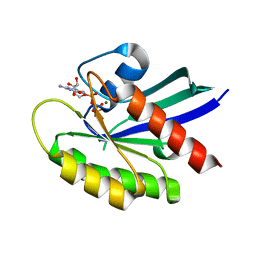 | | Crystal structure of wild-type KRAS4b(1-169) in complex with GMPPNP and Mg ion | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Tran, T.H, Davies, D.R, Edwards, T.E, Simanshu, D.K. | | Deposit date: | 2019-12-20 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Machine learning-driven multiscale modeling reveals lipid-dependent dynamics of RAS signaling proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6ML6
 
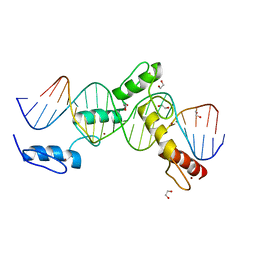 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpA 5mC Modification) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*(5CM)P*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
8E2B
 
 | | N-terminal domain of S. aureus GpsB | | Descriptor: | Cell cycle protein GpsB, GLYCEROL | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2022-08-14 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Staphylococcus aureus FtsZ and PBP4 bind to the conformationally dynamic N-terminal domain of GpsB.
Elife, 13, 2024
|
|
6ML5
 
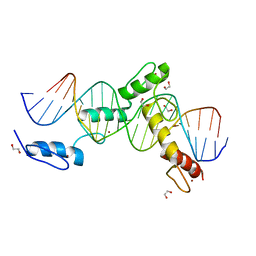 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
8E2C
 
 | |
6ML7
 
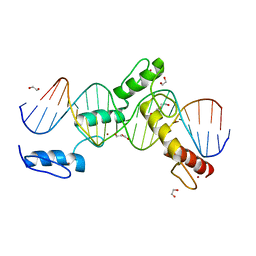 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpG 5mC Modification) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*(5CM)P*GP*AP*AP*TP*T)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
5DEP
 
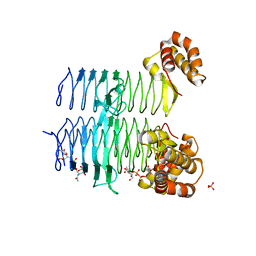 | | Structure of Pseudomonas aeruginosa LpxA in complex with UDP-GlcNAc | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, PHOSPHATE ION, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Smith, E.W, Chen, Y. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structures of Pseudomonas aeruginosa LpxA Reveal the Basis for Its Substrate Selectivity.
Biochemistry, 54, 2015
|
|
5DG3
 
 | |
6UNB
 
 | | Crystal structure of CTX-M-14 in complex with temocillin | | Descriptor: | (2R,4S)-2-[(1S)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-1-methoxy-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4 -carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2019-10-11 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
6UED
 
 | | Apo Pseudomonas aeruginosa LpxD Structure | | Descriptor: | GLYCEROL, MAGNESIUM ION, UDP-3-O-acylglucosamine N-acyltransferase | | Authors: | Chen, Y, Kroeck, K, Sacco, M. | | Deposit date: | 2019-09-20 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
3NMU
 
 | | Crystal Structure of substrate-bound halfmer box C/D RNP | | Descriptor: | 50S ribosomal protein L7Ae, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein, ... | | Authors: | Li, H, Xue, S, Wang, R. | | Deposit date: | 2010-06-22 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.729 Å) | | Cite: | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
2H2H
 
 | |
6ML3
 
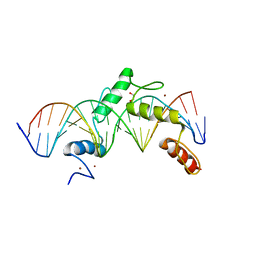 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 2) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*CP*TP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
5VZU
 
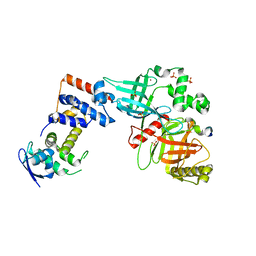 | | Crystal structure of the Skp1-FBXO31-cyclin D1 complex | | Descriptor: | Cyclin D1, F-box only protein 31, PHOSPHATE ION, ... | | Authors: | Li, Y, Jin, K, Hao, B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the phosphorylation-independent recognition of cyclin D1 by the SCFFBXO31 ubiquitin ligase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5IKL
 
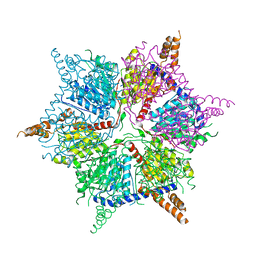 | |
6UUS
 
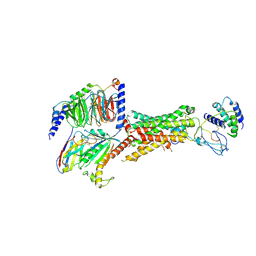 | | CryoEM Structure of the active Adrenomedullin 2 receptor G protein complex with adrenomedullin peptide | | Descriptor: | ADM, Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Sexton, P, Danev, R. | | Deposit date: | 2019-10-31 | | Release date: | 2020-04-01 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure and Dynamics of Adrenomedullin Receptors AM1and AM2Reveal Key Mechanisms in the Control of Receptor Phenotype by Receptor Activity-Modifying Proteins.
Acs Pharmacol Transl Sci, 3, 2020
|
|
2H2G
 
 | |
6UVA
 
 | | CryoEM Structure of the active Adrenomedullin 2 receptor G protein complex with adrenomedullin 2 peptide | | Descriptor: | Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Sexton, P, Danev, R. | | Deposit date: | 2019-11-01 | | Release date: | 2020-04-01 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structure and Dynamics of Adrenomedullin Receptors AM1and AM2Reveal Key Mechanisms in the Control of Receptor Phenotype by Receptor Activity-Modifying Proteins.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6ML2
 
 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 1) | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*CP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ZINC ION, ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
3NBV
 
 | |
4H42
 
 | | Synthesis of a Weak Basic uPA Inhibitor and Crystal Structure of Complex with uPA | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, N-[(2-amino-1,3-benzothiazol-6-yl)carbonyl]glycine, Urokinase-type plasminogen activator | | Authors: | Yu, H.-Y, Gao, D, Zhang, X, Jiang, L.-G, Hong, Z.-B, Yuan, C, Fang, X, Wang, J.-D, Huang, M.-D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Synthesis of a Weak Basic uPA Inhibitor and Crystal Structure of Complex with uPA
CHIN.J.STRUCT.CHEM., 32, 2013
|
|
8F6H
 
 | | Cryo-EM structure of a Zinc-loaded asymmetrical TMD D70A mutant of the YiiP-Fab complex | | Descriptor: | Cadmium and zinc efflux pump FieF, Fab2r heavy chain, Fab2r light chain, ... | | Authors: | Lopez-Redondo, M.L, Hussein, A.K, Stokes, D.L. | | Deposit date: | 2022-11-16 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Energy coupling and stoichiometry of Zn 2+ /H + antiport by the prokaryotic cation diffusion facilitator YiiP.
Elife, 12, 2023
|
|
8F6E
 
 | | Cryo-EM structure of a Zinc-loaded wild-type YiiP-Fab complex | | Descriptor: | Cadmium and zinc efflux pump FieF, Fab heavy chain, Fab light chain, ... | | Authors: | Lopez-Redondo, M.L, Hussein, A.K, Stokes, D.L. | | Deposit date: | 2022-11-16 | | Release date: | 2023-02-08 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Energy coupling and stoichiometry of Zn 2+ /H + antiport by the prokaryotic cation diffusion facilitator YiiP.
Elife, 12, 2023
|
|
