7SRR
 
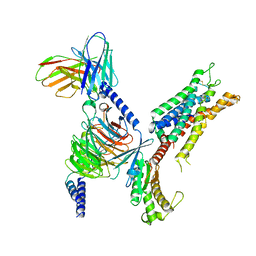 | | 5-HT2B receptor bound to LSD in complex with heterotrimeric mini-Gq protein obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 5-hydroxytryptamine receptor 2B, G protein subunit q (Gi2-mini-Gq chimera), ... | | Authors: | Barros-Alvarez, X, Kim, K, Panova, O, Cao, C, Roth, B.L, Skiniotis, G. | | Deposit date: | 2021-11-08 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Signaling snapshots of a serotonin receptor activated by the prototypical psychedelic LSD.
Neuron, 110, 2022
|
|
7TP3
 
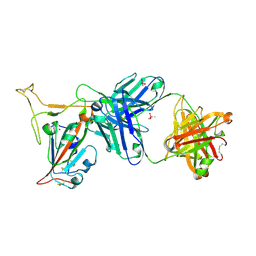 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K288.2 | | Descriptor: | CACODYLATE ION, K288.2 heavy chain, K288.2 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
7TJP
 
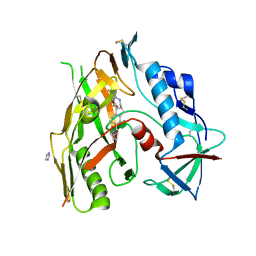 | | HIV-1 gp120 complex with CJF-II-195 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, IMIDAZOLE, ... | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-01-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural and Functional Characterization of Indane-Core CD4-Mimetic Compounds Substituted with Heterocyclic Amines
Acs Medicinal Chemistry Letters, 14, 2023
|
|
7TJO
 
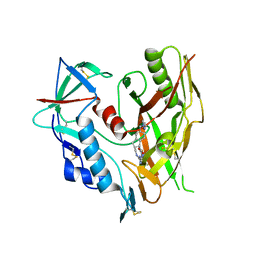 | | HIV-1 gp120 complex with CJF-II-197-S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, IMIDAZOLE, ... | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-01-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structural and Functional Characterization of Indane-Core CD4-Mimetic Compounds Substituted with Heterocyclic Amines
ACS Medicinal Chemistry Letters, 14, 2023
|
|
7TRT
 
 | |
7TRU
 
 | |
7SSW
 
 | |
7SSN
 
 | |
7ST6
 
 | |
7ST7
 
 | |
7ST2
 
 | |
7TI6
 
 | |
7UL2
 
 | | CryoEM Structure of Inactive NTSR1 Bound to SR48692 and Nb6 | | Descriptor: | 2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Nanobody 6, Neurotensin receptor 1, ... | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UL4
 
 | | CryoEM Structure of Inactive MOR Bound to Alvimopan and Mb6 | | Descriptor: | Megabody 6, Mu-type opioid receptor, N-[(2S)-2-{[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethylpiperidin-1-yl]methyl}-3-phenylpropanoyl]glycine | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7U5G
 
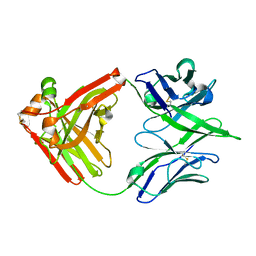 | | ACS122 Fab | | Descriptor: | ACS122 Fab Heavy chain, ACS122 Fab Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-03-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
7UL5
 
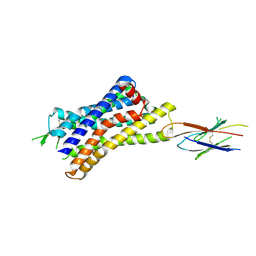 | |
7UL3
 
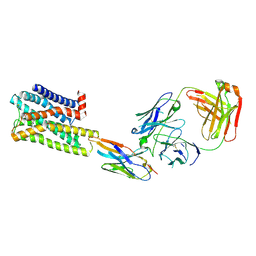 | | CryoEM Structure of Inactive H2R Bound to Famotidine, Nb6M, and NabFab | | Descriptor: | Histamine H2 receptor, NabFab HC, NabFab LC, ... | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TT8
 
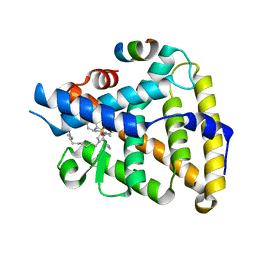 | | Human LRH-1 LBD bound to agonist 6N-10CA and fragment of Tif2 coactivator | | Descriptor: | 10-[(3aR,6S,6aR)-3-phenyl-3a-(1-phenylethenyl)-6-(sulfamoylamino)-1,3a,4,5,6,6a-hexahydropentalen-2-yl]decanoic acid (non-preferred name), Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Cato, M.L, Ortlund, E.A. | | Deposit date: | 2022-01-31 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differential Modulation of Nuclear Receptor LRH-1 through Targeting Buried and Surface Regions of the Binding Pocket.
J.Med.Chem., 65, 2022
|
|
7SSL
 
 | |
7SSO
 
 | |
7SSD
 
 | |
7U8L
 
 | |
7U8M
 
 | |
7U8J
 
 | |
7U0K
 
 | | IOMA class antibody Fab ACS124 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, IOMA Class antibody ACS124 Heavy chain, IOMA Class antibody ACS124 Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
