3DJW
 
 | | The thermo- and acido-stable ORF-99 from the archaeal virus AFV1 | | Descriptor: | ORF99 | | Authors: | Goulet, A, Spinelli, S, Prangishvili, D, van Tilbeurgh, H, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-06-24 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The thermo- and acido-stable ORF-99 from the archaeal virus AFV1
Protein Sci., 18, 2009
|
|
3BCV
 
 | |
6OJ0
 
 | | Cryo-EM reconstruction of Sulfolobus polyhedral virus 1 (SPV1) | | Descriptor: | Structural protein VP4, Uncharacterized protein | | Authors: | Wang, F, Liu, Y, Conway, J.F, Krupovic, M, Prangishvili, D, Egelman, E.H. | | Deposit date: | 2019-04-10 | | Release date: | 2019-10-02 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A packing for A-form DNA in an icosahedral virus.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2UW1
 
 | | Ivy Desaturase Structure | | Descriptor: | (3R)-3-HYDROXY-5,5-DIMETHYLHEXANOIC ACID, FE (III) ION, PLASTID DELTA4 MULTIFUNCTIONAL ACYL-ACYL CARRIER PROTEIN DESATURASE, ... | | Authors: | Guy, J.E, Whittle, E, Kumaran, D, Lindqvist, Y, Shanklin, J. | | Deposit date: | 2007-03-15 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of the Ivy {Delta}4-16:0-Acp Desaturase Reveals Structural Details of the Oxidized Active Site and Potential Determinants of Regioselectivity.
J.Biol.Chem., 282, 2007
|
|
2N7O
 
 | | NMR Structure of Peptide PG-990 in DPC micelles | | Descriptor: | Peptide PG-990 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
6OEY
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor (+)-5-{5-[1-(Pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1-{[(2S)-pyrrolidin-2-yl]methyl}-1H-indole | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1-{[(2S)-pyrrolidin-2-yl]methyl}-1H-indole, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Halgas, O, De Gasparo, R, Harangozo, D, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting a Large Active Site: Structure-Based Design of Nanomolar Inhibitors of Trypanosoma brucei Trypanothione Reductase.
Chemistry, 25, 2019
|
|
6OEX
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor 3-(2-{1-[2-(Piperidin-4-yl)ethyl]-1H-indol-5-yl}-5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3- thiazol-4-yl)-N-(2,2,2-trifluoroethyl)prop-2-yn-1-amine | | Descriptor: | 3-(2-{1-[2-(piperidin-4-yl)ethyl]-1H-indol-5-yl}-5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-4-yl)-N-(2,2,2-trifluoroethyl)prop-2-yn-1-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Halgas, O, De Gasparo, R, Harangozo, D, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting a Large Active Site: Structure-Based Design of Nanomolar Inhibitors of Trypanosoma brucei Trypanothione Reductase.
Chemistry, 25, 2019
|
|
3NQB
 
 | |
2N7T
 
 | | NMR structure of Peptide PG-992 in DPC micelles | | Descriptor: | Peptide PG-992 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
2N7N
 
 | | NMR structure of Peptide PG-989 in DPC micelles | | Descriptor: | Peptide PG-989 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
3P26
 
 | | Crystal structure of S. cerevisiae Hbs1 protein (apo-form), a translational GTPase involved in RNA quality control pathways and interacting with Dom34/Pelota | | Descriptor: | Elongation factor 1 alpha-like protein | | Authors: | van den Elzen, A, Henri, J, Lazar, N, Gas, M.E, Durand, D, Lacroute, F, Nicaise, M, van Tilbeurgh, H, Sraphin, B, Graille, M, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2010-10-01 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2VDU
 
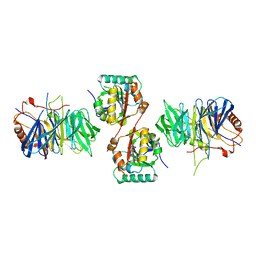 | | Structure of trm8-trm82, THE YEAST TRNA m7G methylation complex | | Descriptor: | PHOSPHATE ION, TRNA (GUANINE-N(7)-)-METHYLTRANSFERASE, TRNA (GUANINE-N(7)-)-METHYLTRANSFERASE-ASSOCIATED WD REPEAT PROTEIN TRM82 | | Authors: | Leulliot, N, Chaillet, M, Durand, D, Ulryck, N, Blondeau, K, Van Tilbeurgh, H. | | Deposit date: | 2007-10-11 | | Release date: | 2007-12-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Yeast tRNA M7G Methylation Complex.
Structure, 16, 2008
|
|
2WB6
 
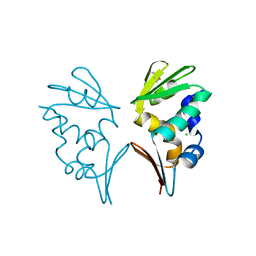 | | Crystal structure of AFV1-102, a protein from the Acidianus Filamentous Virus 1 | | Descriptor: | AFV1-102, CHLORIDE ION | | Authors: | Keller, J, Leulliot, N, Collinet, B, Campanacci, V, Cambillau, C, Pranghisvilli, D, van Tilbeurgh, H. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Afv1-102, a Protein from the Acidianus Filamentous Virus 1.
Protein Sci., 18, 2009
|
|
2W9M
 
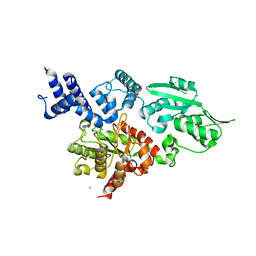 | | Structure of family X DNA polymerase from Deinococcus radiodurans | | Descriptor: | MERCURY (II) ION, POLYMERASE X, ZINC ION | | Authors: | Leulliot, N, Cladiere, L, Lecointe, F, Durand, D, Hubscher, U, van Tilbeurgh, H. | | Deposit date: | 2009-01-27 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The Family X DNA Polymerase from Deinococcus Radioduran Adopts a Non-Standard Extended Conformation.
J.Biol.Chem., 284, 2009
|
|
2OGJ
 
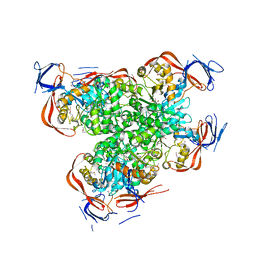 | | Crystal structure of a dihydroorotase | | Descriptor: | Dihydroorotase, IMIDAZOLE, ZINC ION | | Authors: | Sugadev, R, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of a dihydroorotase
To be Published
|
|
2Y7Y
 
 | | APLYSIA CALIFORNICA ACHBP IN APO STATE | | Descriptor: | SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Akdemir, A, Jongejan, A, van Elk, R, Bertrand, S, Perrakis, A, Leurs, R, Smit, A.B, Sixma, T.K, Bertrand, D, De Esch, I.J. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Use of Acetylcholine Binding Protein in the Search for Novel Alpha7 Nicotinic Receptor Ligands. In Silico Docking, Pharmacological Screening, and X-Ray Analysis.
J.Med.Chem., 52, 2009
|
|
2ZBK
 
 | | Crystal structure of an intact type II DNA topoisomerase: insights into DNA transfer mechanisms | | Descriptor: | RADICICOL, Type 2 DNA topoisomerase 6 subunit B, Type II DNA topoisomerase VI subunit A | | Authors: | Graille, M, Cladiere, L, Durand, D, Lecointe, F, Forterre, P, van Tilbeurgh, H, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2007-10-22 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Crystal Structure of an Intact Type II DNA Topoisomerase: Insights into DNA Transfer Mechanisms
Structure, 16, 2008
|
|
5BUW
 
 | |
3DF6
 
 | | The thermo- and acido-stable ORF-99 from the archaeal virus AFV1 | | Descriptor: | CALCIUM ION, ORF99 | | Authors: | Goulet, A, Spinelli, S, Prangishvili, D, van Tilbeurgh, H, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-06-11 | | Release date: | 2009-06-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The thermo- and acido-stable ORF-99 from the archaeal virus AFV1
Protein Sci., 18, 2009
|
|
2KC7
 
 | | Solution NMR structure of Bacteroides fragilis protein BF1650. Northeast Structural Genomics Consortium target BfR218 | | Descriptor: | bfr218_protein | | Authors: | Eletsky, A, Wu, Y, Sukumaran, D, Lee, H, Lee, D.Y, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-17 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Bacteroides fragilis protein BF1650. Northeast Structural Genomics Consortium target BfR218
To be Published
|
|
2KCQ
 
 | | Solution structure of protein SRU_2040 from Salinibacter ruber (strain DSM 13855) . Northeast Structural Genomics Consortium target SrR106 | | Descriptor: | Mov34/MPN/PAD-1 family | | Authors: | Wu, Y, Eletsky, A, Zhao, L, Hua, J, Sukumaran, D, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-28 | | Release date: | 2009-02-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein SRU_2040 from Salinibacter ruber (strain DSM 13855). Northeast Structural Genomics Consortium target SrR106
To be Published
|
|
3S2K
 
 | | Structural basis of Wnt signaling inhibition by Dickkopf binding to LRP5/6. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ahn, V.E, Chu, M.L.-H, Choi, H.-J, Tran, D, Abo, A, Weis, W.I. | | Deposit date: | 2011-05-16 | | Release date: | 2011-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Wnt Signaling Inhibition by Dickkopf Binding to LRP5/6.
Dev.Cell, 21, 2011
|
|
2MMZ
 
 | | Solution structure of the apo form of human glutaredoxin 5 | | Descriptor: | Glutaredoxin-related protein 5, mitochondrial | | Authors: | Banci, L, Brancaccio, D, Ciofi-Baffoni, S, Del Conte, R, Gadepalli, R, Mikolajczyk, M, Neri, S, Piccioli, M, Winkelmann, J. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | [2Fe-2S] cluster transfer in iron-sulfur protein biogenesis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2KKS
 
 | | Solution Structure Of Protein DSY2949 From Desulfitobacterium hafniense. Northeast Structural Genomics Consortium Target DhR27 | | Descriptor: | Uncharacterized protein | | Authors: | Wu, Y, Mills, J.L, Wang, H, Ciccosanti, C, Jiang, M, Sukumaran, D, Zhang, Q, Nair, R, Rost, B, Acton, T, Xiao, R, Swapna, G.V.T, Everett, J, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Of Protein DSY2949 From Desulfitobacterium hafniense. Northeast Structural Genomics Consortium Target DhR27
To be Published
|
|
6PCW
 
 | | Human PIM1 bound to benzothiophene inhibitor 213 | | Descriptor: | 4-[5-(cyclopropylcarbamoyl)thiophen-2-yl]-1-benzothiophene-2-carboxamide, GLYCEROL, Peptide, ... | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-18 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PIM1 bound to benzothiophene inhibitor
To Be Published
|
|
