4WTC
 
 | |
4WTE
 
 | |
4WTG
 
 | |
4WTI
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-ACGG, RNA PRIMER 5'-PCC, MN2+, AND GDP | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTK
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-AGCC, RNA PRIMER 5'-PGG, MN2+, AND CDP | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTL
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-UACC, RNA PRIMER 5'-PGG, MN2+, AND UDP | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
8JYS
 
 | |
6FRK
 
 | | Structure of a prehandover mammalian ribosomal SRP and SRP receptor targeting complex | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kobayashi, K, Jomaa, A, Ban, N. | | Deposit date: | 2018-02-16 | | Release date: | 2018-03-28 | | Last modified: | 2018-05-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a prehandover mammalian ribosomal SRP·SRP receptor targeting complex.
Science, 360, 2018
|
|
4WZ6
 
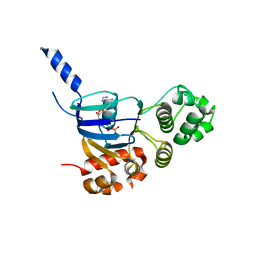 | | Human CFTR aa389-678 (NBD1), deltaF508 with three solubilizing mutations, bound ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Byrnes, L.J, Hall, J. | | Deposit date: | 2014-11-18 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Binding screen for cystic fibrosis transmembrane conductance regulator correctors finds new chemical matter and yields insights into cystic fibrosis therapeutic strategy.
Protein Sci., 25, 2016
|
|
6MSJ
 
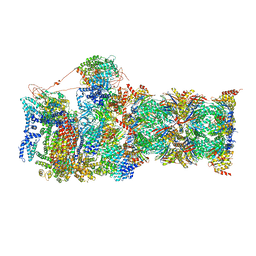 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSG
 
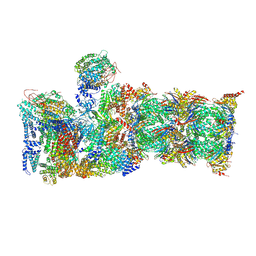 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
7OXL
 
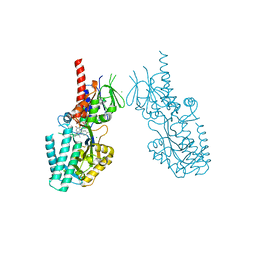 | | Crystal structure of human Spermine Oxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FAD-MDL72527 adduct, ... | | Authors: | Impagliazzo, A, Johannsson, S, Thomsen, M, Krapp, S. | | Deposit date: | 2021-06-22 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human spermine oxidase in complex with a highly selective allosteric inhibitor.
Commun Biol, 5, 2022
|
|
7OY0
 
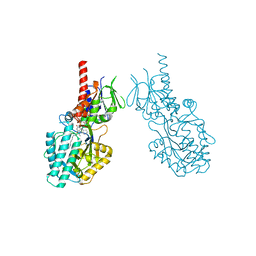 | | Structure of human Spermine Oxidase in complex with a highly selective allosteric inhibitor | | Descriptor: | 4-[(4-imidazo[1,2-a]pyridin-3-yl-1,3-thiazol-2-yl)amino]phenol, CHLORIDE ION, FAD-MDL72527 adduct, ... | | Authors: | Impagliazzo, A, Thomsen, M, Johannsson, S, Krapp, S. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of human spermine oxidase in complex with a highly selective allosteric inhibitor.
Commun Biol, 5, 2022
|
|
6MSE
 
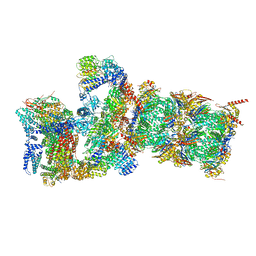 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSB
 
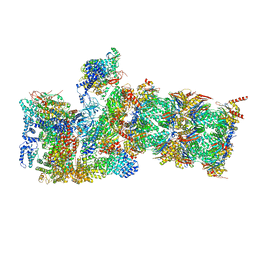 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSH
 
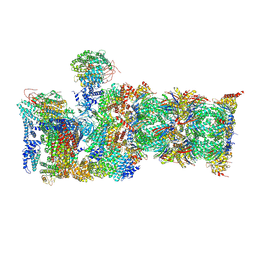 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6OAZ
 
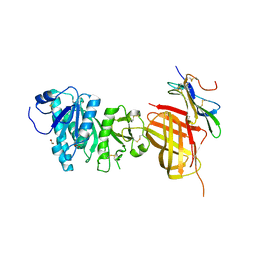 | | Apo Structure of WT Lipoprotein Lipase in Complex with GPIHBP1 Mutant N78D N82D produced in HEK293-F cells | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Arora, R, Horton, P.A, Benson, T.E, Romanowski, M.J. | | Deposit date: | 2019-03-19 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of lipoprotein lipase in complex with GPIHBP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6MSD
 
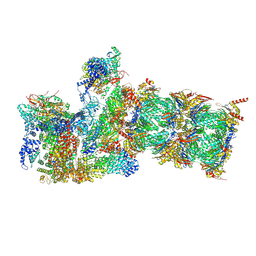 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6NDX
 
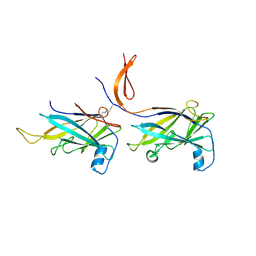 | |
6NAW
 
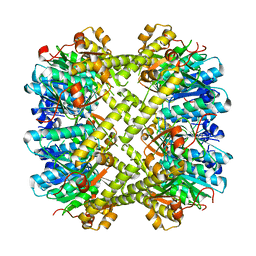 | |
6NAQ
 
 | | Crystal structure of Neisseria meningitidis ClpP protease in Apo form | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, POTASSIUM ION | | Authors: | Houry, W.A, Mabanglo, M.F, Pai, E.F, Eger, B.T, Bryson, S. | | Deposit date: | 2018-12-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores.
Commun Biol, 2, 2019
|
|
8SAI
 
 | | Cryo-EM structure of GPR34-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Yong, X.H, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2023-04-01 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6R9W
 
 | | Crystal structure of InhA in complex with AP-124 inhibitor | | Descriptor: | (2~{S})-1-(benzimidazol-1-yl)-3-(2,3-dihydro-1~{H}-inden-5-yloxy)propan-2-ol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Takebayashi, Y, Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-04-04 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of New and Potent InhA Inhibitors as Antituberculosis Agents: Structure-Based Virtual Screening Validated by Biological Assays and X-ray Crystallography.
J.Chem.Inf.Model., 60, 2020
|
|
6NDT
 
 | |
6NB1
 
 | | Crystal structure of Escherichia coli ClpP protease complexed with small molecule activator, ACP1-06 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, GLYCEROL, N-{2-[(2-chlorophenyl)sulfanyl]ethyl}-2-methyl-2-{[5-(trifluoromethyl)pyridin-2-yl]sulfonyl}propanamide | | Authors: | Mabanglo, M.F, Houry, W.A, Eger, B.T, Bryson, S, Pai, E.F. | | Deposit date: | 2018-12-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores.
Commun Biol, 2, 2019
|
|
