7E88
 
 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7WGC
 
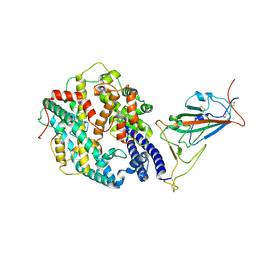 | | Neutral Omicron Spike Trimer in complex with ACE2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Cui, Z. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-22 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7WG7
 
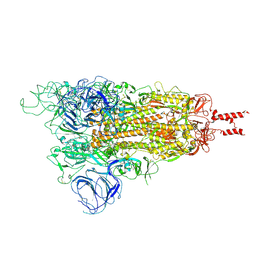 | | Acidic Omicron Spike Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Cui, Z. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E86
 
 | |
7WG9
 
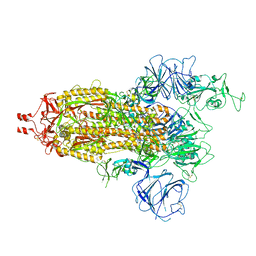 | | Delta Spike Trimer(1 RBD Up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cui, Z. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7WGB
 
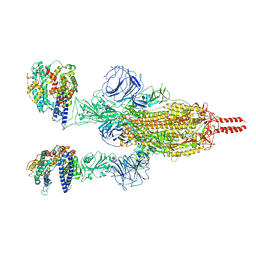 | | Neutral Omicron Spike Trimer in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Cui, Z. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
4RR5
 
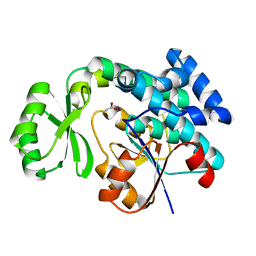 | | The crystal structure of Synechocystis sp. PCC 6803 Malonyl-CoA: ACP Transacylase | | Descriptor: | ACETATE ION, Malonyl CoA-acyl carrier protein transacylase | | Authors: | Liu, Y. | | Deposit date: | 2014-11-06 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.427 Å) | | Cite: | Structural and biochemical characterization of MCAT from photosynthetic microorganism Synechocystis sp. PCC 6803 reveal its stepwise catalytic mechanism
Biochem.Biophys.Res.Commun., 457, 2015
|
|
8W22
 
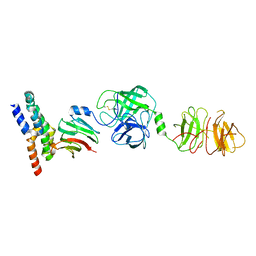 | | Umb1 umbrella toxin particle (local refinement of UmbB1 bound ALF of UmbC1 and UmbA1) | | Descriptor: | Intein C-terminal splicing domain-containing protein, Secreted esterase, Secreted protein | | Authors: | Park, Y.J, Zhao, Q, Seattle Structural Genomics Center for Infectious Disease (SSGCID), DiMaio, F, Mougous, J.D, Veesler, D. | | Deposit date: | 2024-02-19 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Streptomyces umbrella toxin particles block hyphal growth of competing species.
Nature, 629, 2024
|
|
5KPU
 
 | |
7YHK
 
 | | Cryo-EM structure of the HA trimer of A/Beijing/262/1995(H1N1) in complex with neutralizing antibody 12H5 | | Descriptor: | 12H5 heavy chain, 12H5 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, Q, Li, S, Li, T, Xue, W, Sun, H. | | Deposit date: | 2022-07-13 | | Release date: | 2022-08-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
7XUX
 
 | |
7N61
 
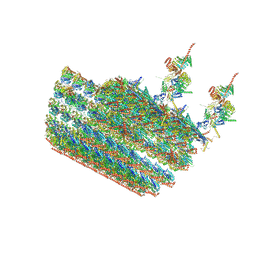 | | structure of C2 projections and MIPs | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FAP147, FAP178, ... | | Authors: | Han, L, Zhang, K. | | Deposit date: | 2021-06-07 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of an active central apparatus.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7N6G
 
 | | C1 of central pair | | Descriptor: | CPC1, Calmodulin, DPY30, ... | | Authors: | Han, L, Zhang, K. | | Deposit date: | 2021-06-08 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of an active central apparatus.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5XHS
 
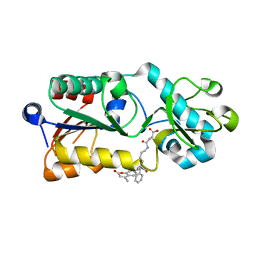 | | Crystal structure of SIRT5 complexed with a fluorogenic small-molecule substrate SuBKA | | Descriptor: | (2S)-2-azanyl-6-[(4-hydroxy-4-oxo-butanoyl)amino]hexanoic acid, 7-AMINO-4-METHYL-CHROMEN-2-ONE, NAD-dependent protein deacylase sirtuin-5, ... | | Authors: | Yu, Y, Li, B, Chen, Q. | | Deposit date: | 2017-04-24 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Interactions between sirtuins and fluorogenic small-molecule substrates offer insights into inhibitor design
Rsc Adv, 7, 2017
|
|
2KLK
 
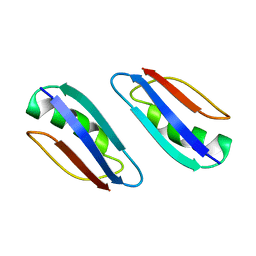 | | Solution structure of GB1 A34F mutant with RDC and SAXS | | Descriptor: | IMMUNOGLOBULIN G-BINDING PROTEIN G | | Authors: | Wang, J, Zuo, X, Yu, P, Byeon, I.L, Jung, J, Schwieters, C.D, Gronenborn, A.M, Wang, Y. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Determination of multicomponent protein structures in solution using global orientation and shape restraints.
J.Am.Chem.Soc., 131, 2009
|
|
8WM4
 
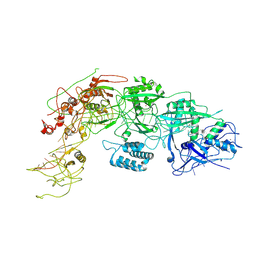 | | Cryo-EM structure of DiCas7-11 in complex with crRNA | | Descriptor: | CRISPR-associated RAMP family protein, ZINC ION, crRNA (38-MER) | | Authors: | Ma, H.Y, Tang, X.D. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of negative regulation of CRISPR-Cas7-11 by TPR-CHAT.
Signal Transduct Target Ther, 9, 2024
|
|
8WML
 
 | |
8WMC
 
 | |
8WMI
 
 | |
5EOF
 
 | | Crystal structure of OPTN NTD and TBK1 CTD complex | | Descriptor: | Optineurin, Serine/threonine-protein kinase TBK1 | | Authors: | Li, F, Xie, X, Liu, J, Pan, L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
5EP6
 
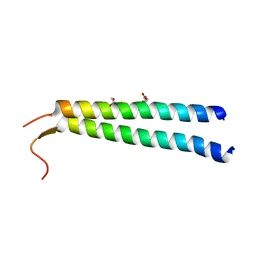 | | The crystal structure of NAP1 in complex with TBK1 | | Descriptor: | 5-azacytidine-induced protein 2, GLYCEROL, Serine/threonine-protein kinase TBK1 | | Authors: | Li, F, Xie, X, Liu, J, Pan, L. | | Deposit date: | 2015-11-11 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
6UBI
 
 | |
7WHZ
 
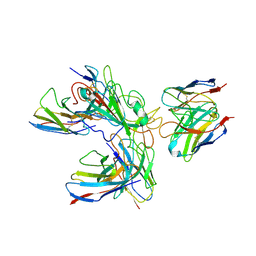 | | SARS-CoV-2 spike protein in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
