4XIQ
 
 | | Discovery of novel oxazepine and diazepine carboxamides as two new classes of heat shock protein 90 inhibitors | | Descriptor: | 8,11,11-trimethyl-9-oxo-6,7,9,10,11,12-hexahydroindolo[2,1-d][1,5]benzoxazepine-3-carboxamide, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Neubert, T, Zuccola, H.J. | | Deposit date: | 2015-01-07 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of novel oxazepine and diazepine carboxamides as two new classes of heat shock protein 90 inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4XIT
 
 | | Discovery of novel oxazepine and diazepine carboxamides as two new classes of heat shock protein 90 inhibitors | | Descriptor: | 2-chloro-5-(furan-2-ylmethyl)-8,11,11-trimethyl-9-oxo-6,7,9,10,11,12-hexahydro-5H-indolo[1,2-a][1,5]benzodiazepine-3-carboxamide, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Zuccola, H.J, Neubert, T. | | Deposit date: | 2015-01-07 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of novel oxazepine and diazepine carboxamides as two new classes of heat shock protein 90 inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5VQM
 
 | | Clostridium difficile TcdB-GTD bound to PA41 Fab | | Descriptor: | PA41 FAB HEAVY CHAIN, PA41 FAB LIGHT CHAIN, Toxin B | | Authors: | Kroh, H.K, Spiller, B.W, Lacy, D.B. | | Deposit date: | 2017-05-09 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A neutralizing antibody that blocks delivery of the enzymatic cargo of Clostridium difficile toxin TcdB into host cells.
J. Biol. Chem., 293, 2018
|
|
6CYF
 
 | | PcrV fragment with bound Fab | | Descriptor: | IgG1 antibody, heavy chain fragment, kappa light chain, ... | | Authors: | Oganesyan, V. | | Deposit date: | 2018-04-05 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Pseudomonas aeruginosa PcrV and Psl, the Molecular Targets of Bispecific Antibody MEDI3902, Are Conserved Among Diverse Global Clinical Isolates.
J. Infect. Dis., 218, 2018
|
|
1GJS
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
1GJT
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
2UXZ
 
 | | Two-Carbon-Elongated HIV-1 Protease Inhibitors with a Tertiary- Alcohol-Containing Transition-State Mimic | | Descriptor: | HIV-1 PROTEASE, METHYL [(1S)-1-({2-[(4R)-4-BENZYL-4-HYDROXY-5-{[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]AMINO}-5-OXOPENTYL]-2-(4-BROMOBENZYL)HYDRAZINO}CARBONYL)-2,2-DIMETHYLPROPYL]CARBAMATE | | Authors: | Ginman, N, Samuelsson, B, Hallberg, A, Unge, J.T, Unge, T.K. | | Deposit date: | 2007-04-02 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Two-Carbon-Elongated HIV-1 Protease Inhibitors with a Tertiary-Alcohol-Containing Transition-State Mimic.
J.Med.Chem., 51, 2008
|
|
2UY0
 
 | | Two-Carbon-Elongated HIV-1 Protease Inhibitors with a Tertiary- Alcohol-Containing Transition-State Mimic | | Descriptor: | HIV-1 PROTEASE, METHYL [(1S)-1-({2-[(4R)-4-BENZYL-4-HYDROXY-5-{[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]AMINO}-5-OXOPENTANOYL]-2-(4-BROMOBENZYL)HYDRAZINO}CARBONYL)-2,2-DIMETHYLPROPYL]CARBAMATE | | Authors: | Ginman, N, Samuelsson, B, Hallberg, A, Unge, J.T, Unge, T.K. | | Deposit date: | 2007-04-02 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Two-Carbon-Elongated HIV-1 Protease Inhibitors with a Tertiary-Alcohol-Containing Transition-State Mimic.
J.Med.Chem., 51, 2008
|
|
2XN2
 
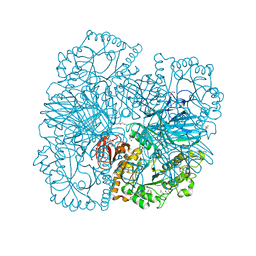 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with galactose | | Descriptor: | ALPHA-GALACTOSIDASE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
4PNJ
 
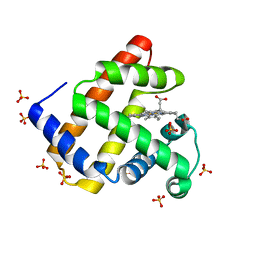 | | Recombinant Sperm Whale P6 Myoglobin Solved with Single Pulse Free Electron Laser Data | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Cohen, A, Gonzalez, A, Lam, W, Lyubimov, A, Sauter, N, Tsai, Y, Uervirojnangkoorn, M, Brunger, A, Soltis, M. | | Deposit date: | 2014-05-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Goniometer-based femtosecond crystallography with X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2C52
 
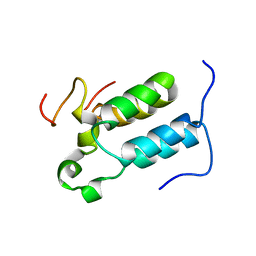 | | Structural diversity in CBP p160 complexes | | Descriptor: | CREB-BINDING PROTEIN, NUCLEAR RECEPTOR COACTIVATOR 1 | | Authors: | Waters, L.C, Yue, B, Veverka, V, Renshaw, P.S, Bramham, J, Matsuda, S, Frenkiel, T, Kelly, G, Muskett, F.W, Carr, M.D, Heery, D.M. | | Deposit date: | 2005-10-25 | | Release date: | 2006-03-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in p160/CREB-binding protein coactivator complexes.
J. Biol. Chem., 281, 2006
|
|
2XN1
 
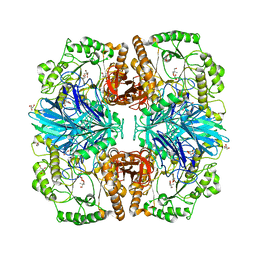 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-GALACTOSIDASE, GLYCEROL | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
2XN0
 
 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM, PtCl4 derivative | | Descriptor: | ALPHA-GALACTOSIDASE, GLYCEROL, PLATINUM (II) ION | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
2J23
 
 | | Cross-reactivity and crystal structure of Malassezia sympodialis Thioredoxin (Mala s 13), a member of a new pan-allergen family | | Descriptor: | THIOREDOXIN | | Authors: | Limacher, A, Glaser, A.G, Meier, C, Scapozza, L, Crameri, R. | | Deposit date: | 2006-08-16 | | Release date: | 2006-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Cross-reactivity and 1.4-A crystal structure of Malassezia sympodialis thioredoxin (Mala s 13), a member of a new pan-allergen family.
J Immunol., 178, 2007
|
|
2X7A
 
 | | Structural basis of HIV-1 tethering to membranes by the Bst2-tetherin ectodomain | | Descriptor: | BONE MARROW STROMAL ANTIGEN 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Natrajan, G, McCarthy, A.A, Weissenhorn, W. | | Deposit date: | 2010-02-25 | | Release date: | 2010-04-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural Basis of HIV-1 Tethering to Membranes by the Bst-2/Tetherin Ectodomain.
Cell Host Microbe, 7, 2010
|
|
5T1Y
 
 | | MLA10 coiled-coil fragment | | Descriptor: | MLA10, TRIETHYLENE GLYCOL | | Authors: | Williams, S.J, Kobe, B, Bentham, A, Ericsson, D.J. | | Deposit date: | 2016-08-22 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The CC domain structure from the wheat stem rust resistance protein Sr33 challenges paradigms for dimerization in plant NLR proteins.
Proc.Natl.Acad.Sci.USA, 2016
|
|
5TCI
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - BRD4592-bound form | | Descriptor: | (2R,3S,4R)-3-(2'-fluoro[1,1'-biphenyl]-4-yl)-4-(hydroxymethyl)azetidine-2-carbonitrile, FORMIC ACID, MALONATE ION, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Wellington, S, Nag, P.P, Fisher, S.L, Schreiber, S.L, Hung, D.T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
5TCH
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - ligand-free form, TrpA-G66V mutant | | Descriptor: | FORMIC ACID, MALONATE ION, Tryptophan synthase alpha chain, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Wellington, S, Nag, P.P, Fisher, S.L, Schreiber, S.L, Hung, D.T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
5TCJ
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - aminoacrylate and BRD4592-bound form | | Descriptor: | (2R,3S,4R)-3-(2'-fluoro[1,1'-biphenyl]-4-yl)-4-(hydroxymethyl)azetidine-2-carbonitrile, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CESIUM ION, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Wellington, S, Nag, P.P, Fisher, S.L, Schreiber, S.L, Hung, D.T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
5TCG
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - aminoacrylate-bound form | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CESIUM ION, FORMIC ACID, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
5TCF
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - ligand-free form | | Descriptor: | FORMIC ACID, MALONATE ION, POTASSIUM ION, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
