7RN0
 
 | |
7RN1
 
 | |
8IEX
 
 | | Solution structure of AtWRKY11-DBD | | Descriptor: | Probable WRKY transcription factor 11, ZINC ION | | Authors: | Dong, X, Hu, Y.F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of Arabidopsis transcription factor WRKY11.
Biochem.Biophys.Res.Commun., 653, 2023
|
|
5KZF
 
 | |
5DIK
 
 | | Crystal structure of apo-lpg0406, a carboxymuconolactone decarboxylase family protein from Legionella pneumophila | | Descriptor: | Alkyl hydroperoxide reductase AhpD | | Authors: | Chen, X, Gong, X, Zhang, N, Ge, H. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of lpg0406, a carboxymuconolactone decarboxylase family protein possibly involved in antioxidative response from Legionella pneumophila
Protein Sci., 24, 2015
|
|
5KWA
 
 | |
6WTT
 
 | | Crystals Structure of the SARS-CoV-2 (COVID-19) main protease with inhibitor GC-376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Sacco, M, Ma, C, Chen, Y, Wang, J. | | Deposit date: | 2020-05-03 | | Release date: | 2020-05-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease.
Cell Res., 30, 2020
|
|
6XOG
 
 | | Structure of SUMO1-ML786519 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOH
 
 | | Structure of SUMO1-ML00789344 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOI
 
 | | Structure of SUMO1-ML00752641 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
4HP3
 
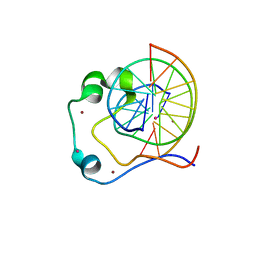 | | Crystal structure of Tet3 in complex with a CpG dsDNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*CP*GP*TP*TP*GP*GP*C)-3'), LOC100036628 protein, UNKNOWN ATOM OR ION, ... | | Authors: | Chao, X, Tempel, W, Bian, C, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-10-23 | | Release date: | 2012-12-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tet3 CXXC Domain and Dioxygenase Activity Cooperatively Regulate Key Genes for Xenopus Eye and Neural Development.
Cell(Cambridge,Mass.), 151, 2012
|
|
4HP1
 
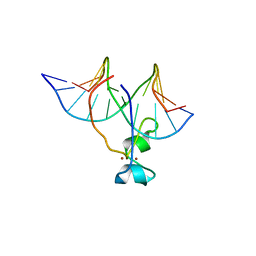 | | Crystal structure of Tet3 in complex with a non-CpG dsDNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*CP*(5CM)P*GP*GP*TP*GP*GP*C)-3'), LOC100036628 protein, ZINC ION | | Authors: | Chao, X, Tempel, W, Bian, C, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-10-23 | | Release date: | 2012-12-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tet3 CXXC Domain and Dioxygenase Activity Cooperatively Regulate Key Genes for Xenopus Eye and Neural Development.
Cell(Cambridge,Mass.), 151, 2012
|
|
5JDK
 
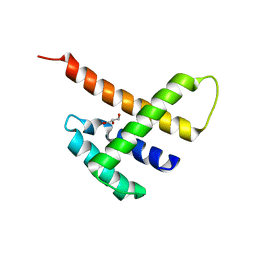 | |
9LON
 
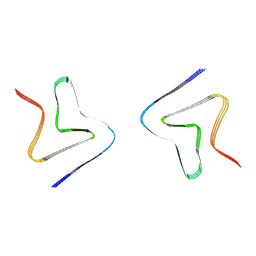 | | Cryo-EM structure of alpha-synuclein P8 fibril | | Descriptor: | Alpha-synuclein | | Authors: | Xia, W.C, Liu, C. | | Deposit date: | 2025-01-23 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Fibril fuzzy coat is important for alpha-synuclein pathological transmission activity.
Neuron, 113, 2025
|
|
7RK0
 
 | | Crystal structure of Thermovibrio ammonificans THI4 | | Descriptor: | 2-[(E)-[(4R)-5-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-4-oxidanyl-3-oxidanylidene-pentan-2-ylidene]amino]ethanoic acid, FE (III) ION, Thiamine thiazole synthase | | Authors: | Li, Q, Bruner, S.D. | | Deposit date: | 2021-07-21 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure and function of aerotolerant, multiple-turnover THI4 thiazole synthases.
Biochem.J., 478, 2021
|
|
7KVZ
 
 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN-2 | | Descriptor: | (2R,5R,7R,8R,10S,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2,10,16-trihydroxy-14-[(pyrimidin-4-yl)oxy]decahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R.J. | | Deposit date: | 2020-11-29 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
7KW1
 
 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN-3 | | Descriptor: | (2R,5R,7R,8R,10R,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-16-hydroxy-14-[(pyrimidin-4-yl)oxy]-2,10-disulfanyldecahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R. | | Deposit date: | 2020-11-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
7KVX
 
 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN 1 | | Descriptor: | (2R,5R,7R,8R,10R,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-16-hydroxy-14-[(pyrimidin-4-yl)amino]-2,10-disulfanyldecahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R. | | Deposit date: | 2020-11-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
6XFN
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW243 | | Descriptor: | 3C-like proteinase, GLYCEROL, UAW243 | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-15 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XBI
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW248 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-06 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XH7
 
 | | CueR-TAC without RNA | | Descriptor: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
6XH8
 
 | | CueR-transcription activation complex with RNA transcript | | Descriptor: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
6XBH
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW247 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-06 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XBG
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW246 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XA4
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW241 | | Descriptor: | 3C-like proteinase, GLYCEROL, inhibitor UAW241 | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
