5JLG
 
 | | The X-ray structure of the adduct formed in the reaction between bovine pancreatic ribonuclease and compound I, a piano-stool organometallic Ru(II) arene compound containing an O,S-chelating ligand | | Descriptor: | DIMETHYL SULFOXIDE, RUTHENIUM ION, Ribonuclease pancreatic, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2016-04-27 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Unusual mode of protein binding by a cytotoxic pi-arene ruthenium(ii) piano-stool compound containing an O,S-chelating ligand.
Dalton Trans, 45, 2016
|
|
8A5N
 
 | |
8BOV
 
 | | X-ray structure of the adduct formed upon reaction of the five-coordinate Pt(II) complex, 1-Me,Me, with HEWL at pH 7.5 | | Descriptor: | 1-[1,3-dimethyl-4-(1~{H}-1,2,3-triazol-5-yl)imidazol-1-ium-2-yl]-1,2',11'-trimethyl-spiro[1$l^{6}-platinacycloprop-2-ene-1,15'-1,12-diaza-15$l^{6}-platinatetracyclo[10.2.1.0^{5,14}.0^{8,13}]pentadeca-2,4,6,8,10,13-hexaene], 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Ferraro, G, Tito, G, Merlino, A. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Impact of Hydrophobic Chains in Five-Coordinate Glucoconjugate Pt(II) Anticancer Agents.
Int J Mol Sci, 24, 2023
|
|
8BOY
 
 | | X-ray structure of the adduct formed upon reaction of the five-coordinate Pt(II) complex, 1-Me,Me, with HEWL at pH 4.0 | | Descriptor: | 1-[1,3-dimethyl-4-(1~{H}-1,2,3-triazol-5-yl)imidazol-1-ium-2-yl]-1,2',11'-trimethyl-spiro[1$l^{6}-platinacycloprop-2-ene-1,15'-1,12-diaza-15$l^{6}-platinatetracyclo[10.2.1.0^{5,14}.0^{8,13}]pentadeca-2,4,6,8,10,13-hexaene], Lysozyme C, NITRATE ION, ... | | Authors: | Ferraro, G, Tito, G, Merlino, A. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Impact of Hydrophobic Chains in Five-Coordinate Glucoconjugate Pt(II) Anticancer Agents.
Int J Mol Sci, 24, 2023
|
|
4J1B
 
 | |
6ENV
 
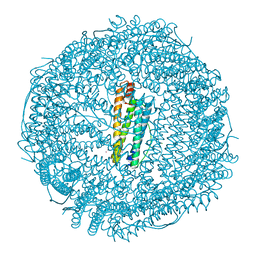 | |
4KXH
 
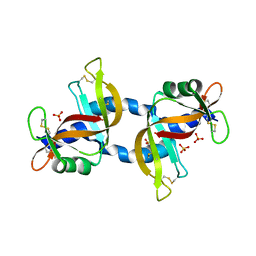 | | The X-ray crystal structure of a dimeric variant of human pancreatic ribonuclease | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SODIUM ION, ... | | Authors: | Pica, A, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2013-05-26 | | Release date: | 2013-10-02 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional domain swapping and supramolecular protein assembly: insights from the X-ray structure of a dimeric swapped variant of human pancreatic RNase.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4L55
 
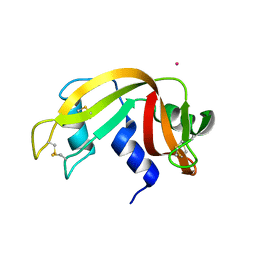 | |
3GQG
 
 | | Crystal structure at acidic pH of the ferric form of the Root effect hemoglobin from Trematomus bernacchii. | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vergara, A, Franzese, M, Merlino, A, Bonomi, G, Mazzarella, L. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Correlation between hemichrome stability and the root effect in tetrameric hemoglobins.
Biophys.J., 97, 2009
|
|
4IAU
 
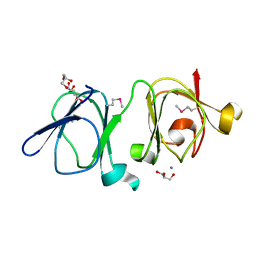 | | Atomic resolution structure of Geodin, a beta-gamma crystallin from Geodia cydonium | | Descriptor: | Beta-gamma-crystallin, CALCIUM ION, GLYCEROL | | Authors: | Vergara, A, Grassi, M, Sica, F, Mazzarella, L, Merlino, A. | | Deposit date: | 2012-12-07 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | A novel interdomain interface in crystallins: structural characterization of the [beta][gamma]-crystallin from Geodia cydonium at 0.99 A resolution
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4LFX
 
 | |
4LGK
 
 | |
4LFP
 
 | |
9EX2
 
 | | X-ray structure of a polyoxidovanadate/lysozyme adduct obtained when the protein is treated with [VIVO(acac)2] in 1.1 M NaCl, 0.1 M sodium acetate at pH 4.0 (Structure C) | | Descriptor: | CHLORIDE ION, Lysozyme C, Polyoxidovanadate complex, ... | | Authors: | Tito, G, Merlino, A, Ferraro, G. | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.172 Å) | | Cite: | Non-Covalent and Covalent Binding of New Mixed-Valence Cage-like Polyoxidovanadate Clusters to Lysozyme.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
9EX0
 
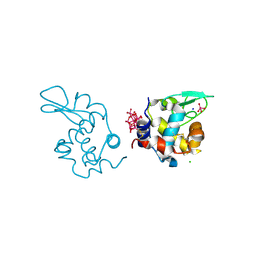 | | X-ray structure of a polyoxidovanadate/lysozyme adduct obtained when the protein is treated with [VIVO(acac)2] in 1.1 M NaCl, 0.1 M sodium acetate at pH 4.0 (Structure A) | | Descriptor: | CHLORIDE ION, Lysozyme C, Polyoxidovanadate complex, ... | | Authors: | Tito, G, Merlino, A, Ferraro, G. | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Non-Covalent and Covalent Binding of New Mixed-Valence Cage-like Polyoxidovanadate Clusters to Lysozyme.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
9EX1
 
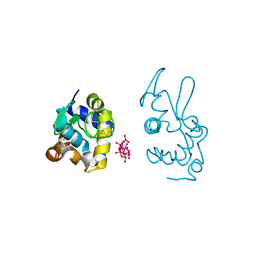 | | X-ray structure of a polyoxidovanadate/lysozyme adduct obtained when the protein is treated with [VIVO(acac)2] in 1.1 M NaCl, 0.1 M sodium acetate at pH 4.0 (Structure B) | | Descriptor: | CHLORIDE ION, Lysozyme C, Polyoxidovanadate complex | | Authors: | Tito, G, Merlino, A, Ferraro, G. | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Non-Covalent and Covalent Binding of New Mixed-Valence Cage-like Polyoxidovanadate Clusters to Lysozyme.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
4QH3
 
 | |
4QGZ
 
 | |
4QY9
 
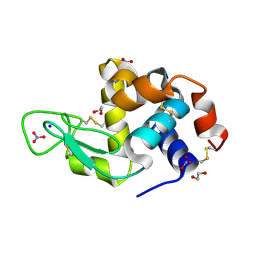 | | X-ray structure of the adduct between hen egg white lysozyme and Auoxo3, a cytotoxic gold(III) compound | | Descriptor: | 1,2-ETHANEDIOL, GOLD ION, Lysozyme C, ... | | Authors: | Russo Krauss, I, Merlino, A. | | Deposit date: | 2014-07-24 | | Release date: | 2014-11-05 | | Last modified: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Interactions of gold-based drugs with proteins: the structure and stability of the adduct formed in the reaction between lysozyme and the cytotoxic gold(iii) compound Auoxo3.
Dalton Trans, 43, 2014
|
|
8PFU
 
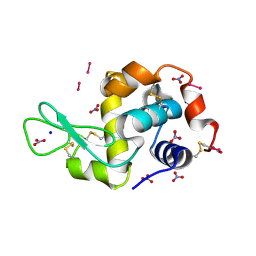 | | X-ray structure of the adduct formed upon reaction of Lysozyme with K3[Ru2(CO3)4] in condition A | | Descriptor: | 6,8-bis(4-fluorophenyl)-1,5-bis(oxidanyl)-2,4-dioxa-6,8-diaza-1$l^{4},5$l^{4}-diruthenabicyclo[3.3.0]octan-3-one, CARBONATE ION, Lysozyme C, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Charge effect in protein metalation reactions by diruthenium complexes
Inorg Chem Front, 2023
|
|
8PFV
 
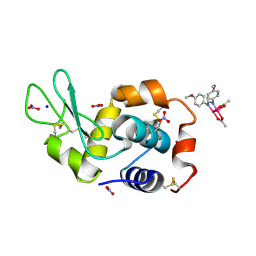 | | X-ray structure of the adduct formed upon reaction of Lysozyme with [Ru2Cl(DAniF)(O2CCH3)3] in condition A | | Descriptor: | 9,11-bis(4-methoxyphenyl)-3,7-dimethyl-2,4,6,8-tetraoxa-9,11-diaza-1$l^{4},5$l^{4}-diruthenatricyclo[3.3.3.0^{1,5}]undecane, CHLORIDE ION, Lysozyme C, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Charge effect in protein metalation reactions by diruthenium complexes
Inorg Chem Front, 2023
|
|
8PFY
 
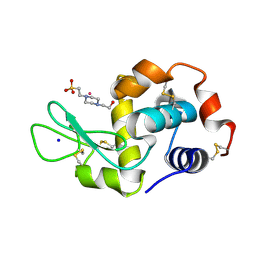 | |
8PFT
 
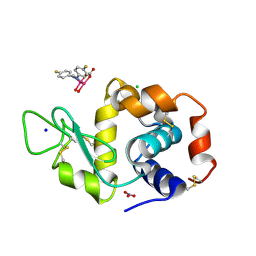 | | X-ray structure of the adduct formed upon reaction of Lysozyme with K2[Ru2(D-p-FPhF)(CO3)3] in condition A | | Descriptor: | 6,8-bis(4-fluorophenyl)-1,5-bis(oxidanyl)-2,4-dioxa-6,8-diaza-1$l^{4},5$l^{4}-diruthenabicyclo[3.3.0]octan-3-one, CHLORIDE ION, Lysozyme C, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Charge effect in protein metalation reactions by diruthenium complexes
Inorg Chem Front, 2023
|
|
8PFW
 
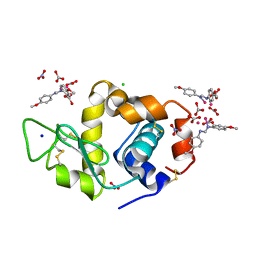 | |
8PFX
 
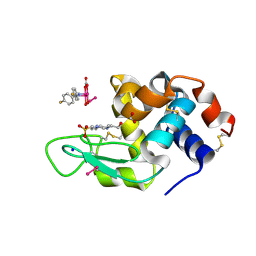 | | X-ray structure of the adduct formed upon reaction of Lysozyme with K2[Ru2(D-p-FPhF)(CO3)3] in condition B | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6,8-bis(4-fluorophenyl)-1,5-bis(oxidanyl)-2,4-dioxa-6,8-diaza-1$l^{4},5$l^{4}-diruthenabicyclo[3.3.0]octan-3-one, FORMIC ACID, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Charge effect in protein metalation reactions by diruthenium complexes
Inorg Chem Front, 2023
|
|
