8WFR
 
 | | The Crystal Structure of PCSK9 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Lu, Y. | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of PCSK9 from Biortus.
To Be Published
|
|
8X2R
 
 | | The Crystal Structure of HSP 90-alpha from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Lu, Y. | | Deposit date: | 2023-11-10 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of HSP 90-alpha from Biortus.
To Be Published
|
|
8X2P
 
 | | The Crystal Structure of LCK from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Lu, Y. | | Deposit date: | 2023-11-10 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of LCK from Biortus.
To Be Published
|
|
8X7V
 
 | | Structure of human SCMC ternary complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,NACHT, LRR and PYD domains-containing protein 5, Oocyte-expressed protein homolog, ... | | Authors: | Chi, P, Ou, G, Liu, S, Lu, Y, Li, J.H, Li, J.L, Wang, X, Deng, D. | | Deposit date: | 2023-11-26 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Cryo-EM structure of the human subcortical maternal complex and the associated discovery of infertility-associated variants.
Nat.Struct.Mol.Biol., 2024
|
|
8X7W
 
 | | Structure of dimeric human SCMC complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,NACHT, LRR and PYD domains-containing protein 5, Oocyte-expressed protein homolog, ... | | Authors: | Chi, P, Ou, G, Liu, S, Lu, Y, Li, J, Li, J, Wang, X, Deng, D. | | Deposit date: | 2023-11-26 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Cryo-EM structure of the human subcortical maternal complex and the associated discovery of infertility-associated variants.
Nat.Struct.Mol.Biol., 2024
|
|
8I0T
 
 | | The cryo-EM structure of human Bact-III complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0R
 
 | | The cryo-EM structure of human Bact-I complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0V
 
 | | The cryo-EM structure of human post-Bact complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0P
 
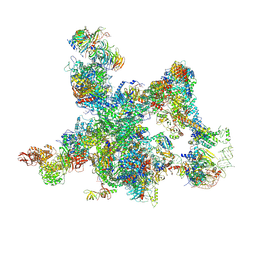 | | The cryo-EM structure of human pre-Bact complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0S
 
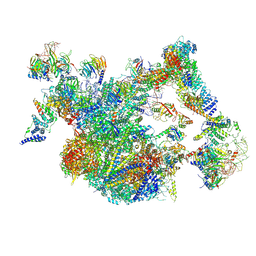 | | The cryo-EM structure of human Bact-II complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0W
 
 | | The cryo-EM structure of human C complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Coiled-coil domain-containing protein 94, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0U
 
 | | The cryo-EM structure of human Bact-IV complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
7ENO
 
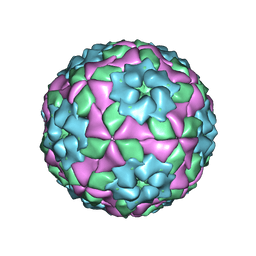 | | Mutant strain M3 of foot-and-mouth disease virus type O | | Descriptor: | VP1 of O type FMDV capsid, VP2 of O type FMDV capsid, VP3 of O type FMDV capsid, ... | | Authors: | Dong, H, Lu, Y. | | Deposit date: | 2021-04-18 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | A Heat-Induced Mutation on VP1 of Foot-and-Mouth Disease Virus Serotype O Enhanced Capsid Stability and Immunogenicity.
J.Virol., 95, 2021
|
|
7ENP
 
 | | wild type of O type Foot-and-mouth disease virus | | Descriptor: | VP1 of O type FMDV capsid protein, VP2 of O type FMDV capsid protein, VP3 of O type FMDV capsid protein, ... | | Authors: | Dong, H, Lu, Y. | | Deposit date: | 2021-04-18 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A Heat-Induced Mutation on VP1 of Foot-and-Mouth Disease Virus Serotype O Enhanced Capsid Stability and Immunogenicity.
J.Virol., 95, 2021
|
|
7W5A
 
 | | The cryo-EM structure of human pre-C*-II complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-06-22 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of exon ligation by human spliceosome.
Mol.Cell, 82, 2022
|
|
7W5B
 
 | | The cryo-EM structure of human C* complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanism of exon ligation by human spliceosome.
Mol.Cell, 82, 2022
|
|
7W59
 
 | | The cryo-EM structure of human pre-C*-I complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of exon ligation by human spliceosome.
Mol.Cell, 82, 2022
|
|
7WSW
 
 | | Cryo-EM structure of the Potassium channel AKT1 from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
7XUF
 
 | | Cryo-EM structure of the AKT1-AtKC1 complex from Arabidopsis thaliana | | Descriptor: | POTASSIUM ION, Potassium channel AKT1, Potassium channel KAT3 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Yang, F, Zhang, Y.M, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
5UK8
 
 | | The co-structure of (R)-4-(6-(1-(cyclopropylsulfonyl)cyclopropyl)-2-(1H-indol-4-yl)pyrimidin-4-yl)-3-methylmorpholine and a rationally designed PI3K-alpha mutant that mimics ATR | | Descriptor: | (R)-4-(6-(1-(cyclopropylsulfonyl)cyclopropyl)-2-(1H-indol-4-yl)pyrimidin-4-yl)-3-methylmorpholine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Mamo, M, Elling, R.A. | | Deposit date: | 2017-01-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rationally Designed PI3K alpha Mutants to Mimic ATR and Their Use to Understand Binding Specificity of ATR Inhibitors.
J. Mol. Biol., 429, 2017
|
|
5UL1
 
 | | The co-structure of 3-amino-6-(4-((1-(dimethylamino)propan-2-yl)sulfonyl)phenyl)-N-phenylpyrazine-2-carboxamide and a rationally designed PI3K-alpha mutant that mimics ATR | | Descriptor: | 3-amino-6-(4-{[(2S)-1-(dimethylamino)propan-2-yl]sulfonyl}phenyl)-N-phenylpyrazine-2-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A, Mamo, M. | | Deposit date: | 2017-01-23 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Rationally Designed PI3K alpha Mutants to Mimic ATR and Their Use to Understand Binding Specificity of ATR Inhibitors.
J. Mol. Biol., 429, 2017
|
|
5UKJ
 
 | | The co-structure of N,N-dimethyl-4-[(6R)-6-methyl-5-(1H-pyrrolo[2,3- b]pyridin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5- a]pyrazin-3-yl]benzenesulfonamide and a rationally designed PI3K-alpha mutant that mimics ATR | | Descriptor: | N,N-dimethyl-4-[(6R)-6-methyl-5-(1H-pyrrolo[2,3-b]pyridin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-3-yl]benzenesulfonamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A, Mamo, M. | | Deposit date: | 2017-01-23 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rationally Designed PI3K alpha Mutants to Mimic ATR and Their Use to Understand Binding Specificity of ATR Inhibitors.
J. Mol. Biol., 429, 2017
|
|
7FCV
 
 | | Cryo-EM structure of the Potassium channel AKT1 mutant from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Zhang, Y.M, Tang, R.F, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
7MVS
 
 | | DNA gyrase complexed with uncleaved DNA and Compound 7 to 2.6A resolution | | Descriptor: | (1S)-2-[(2r,5S)-5-{[(2,3-dihydro-1,4-benzodioxin-6-yl)methyl]amino}-1,3-dioxan-2-yl]-1-(3-fluoro-6-methoxyquinolin-4-yl)ethan-1-ol, CHLORIDE ION, DNA (5'-D(P*AP*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Ratigan, S.C, McElroy, C.A. | | Deposit date: | 2021-05-15 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.60137153 Å) | | Cite: | Optimization of TopoIV Potency, ADMET Properties, and hERG Inhibition of 5-Amino-1,3-dioxane-Linked Novel Bacterial Topoisomerase Inhibitors: Identification of a Lead with In Vivo Efficacy against MRSA.
J.Med.Chem., 64, 2021
|
|
5CMN
 
 | | FLRT3 LRR domain in complex with LPHN3 Olfactomedin domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Latrophilin-3, ... | | Authors: | Lu, Y, Salzman, G, Arac, D. | | Deposit date: | 2015-07-17 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.605 Å) | | Cite: | Structural Basis of Latrophilin-FLRT-UNC5 Interaction in Cell Adhesion.
Structure, 23, 2015
|
|
