6L8D
 
 | |
6L8I
 
 | | Crystal structure of CYP97A3 mutant S290D/W300L/S304V | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Protein LUTEIN DEFICIENT 5, chloroplastic | | Authors: | Niu, G, Guo, Q, Liu, L. | | Deposit date: | 2019-11-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for plant lutein biosynthesis from alpha-carotene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L1H
 
 | |
7O25
 
 | | Complex-B bound [FeFe]-hydrogenase maturase HydE from T. maritima (reaction triggered in the crystal) | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBON MONOXIDE, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O1P
 
 | | [FeFe]-hydrogenase maturase HydE from T. Maritima (C-ter stretch absent) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O26
 
 | | Complex-B bound [FeFe]-hydrogenase maturase HydE fromT. Maritima (5'dA + Methionine) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5'-DEOXYADENOSINE, CARBON MONOXIDE, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O1S
 
 | | Complex-B bound [FeFe]-hydrogenase maturase HydE fromT. Maritima (Wild-type protein) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBON MONOXIDE, CYANIDE ION, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O1T
 
 | | Fe(CO)2CNCl species bound [HydE from T. Maritima | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBON MONOXIDE, CHLORIDE ION, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O1O
 
 | | Complex-B bound [FeFe]-hydrogenase maturase HydE fromT. Maritima (Auxiliary cluster deleted variant) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, CYANIDE ION, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7YQK
 
 | | cryo-EM structure of gammaH2AXK15ub-H4K20me2 nucleosome bound to 53BP1 | | Descriptor: | DNA (145-MER), Histone H2AX, Histone H2B, ... | | Authors: | Ai, H.S, GuoChao, C, Qingyue, G, Ze-Bin, T, Zhiheng, D, Xin, L, Fan, Y, Ziyu, X, Jia-Bin, L, Changlin, T, Liu, L. | | Deposit date: | 2022-08-07 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Chemical Synthesis of Post-Translationally Modified H2AX Reveals Redundancy in Interplay between Histone Phosphorylation, Ubiquitination, and Methylation on the Binding of 53BP1 with Nucleosomes.
J.Am.Chem.Soc., 144, 2022
|
|
7MEY
 
 | | Structure of yeast Ubr1 in complex with Ubc2 and monoubiquitinated N-degron | | Descriptor: | 2-(ethylamino)ethane-1-thiol, E3 ubiquitin-protein ligase UBR1, Monoubiquitinated N-degron, ... | | Authors: | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-11-24 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
7MEX
 
 | | Structure of yeast Ubr1 in complex with Ubc2 and N-degron | | Descriptor: | E3 ubiquitin-protein ligase UBR1, N-degron, Ubiquitin, ... | | Authors: | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
1FQQ
 
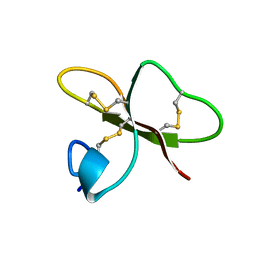 | | SOLUTION STRUCTURE OF HUMAN BETA-DEFENSIN-2 | | Descriptor: | BETA-DEFENSIN-2 | | Authors: | Sawai, M.V, Jia, H.P, Liu, L, Aseyev, V, Wiencek, J.M, McCray Jr, P.B, Ganz, T, Kearney, W.R, Tack, B.F. | | Deposit date: | 2000-09-06 | | Release date: | 2001-04-11 | | Last modified: | 2018-10-24 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of human beta-defensin-2 reveals a novel alpha-helical segment.
Biochemistry, 40, 2001
|
|
8AZT
 
 | | Type II amyloid-beta 42 filaments from high-spin supernatants of aqueous extracts from Alzheimer's disease brains | ABeta42 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Stern, M.A, Meunier, L.A, Liu, W, Cai, Y.Q, Ericsson, M, Liu, L, Selkoe, J.D, Goedert, M, Scheres, H.W.S. | | Deposit date: | 2022-09-06 | | Release date: | 2022-11-02 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Abundant A beta fibrils in ultracentrifugal supernatants of aqueous extracts from Alzheimer's disease brains.
Neuron, 111, 2023
|
|
1G86
 
 | | CHARCOT-LEYDEN CRYSTAL PROTEIN/N-ETHYLMALEIMIDE COMPLEX | | Descriptor: | CHARCOT-LEYDEN CRYSTAL PROTEIN, N-ETHYLMALEIMIDE | | Authors: | Ackerman, S.J, Liu, L, Kwatia, M.A, Savage, M.P, Leonidas, D.D, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2000-11-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Charcot-Leyden crystal protein (galectin-10) is not a dual function galectin with lysophospholipase activity but binds a lysophospholipase inhibitor in a novel structural fashion.
J.Biol.Chem., 277, 2002
|
|
1HDK
 
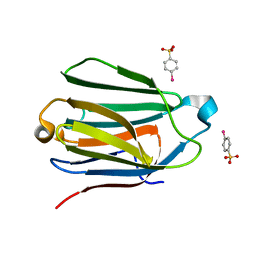 | | Charcot-Leyden Crystal Protein - pCMBS Complex | | Descriptor: | EOSINOPHIL LYSOPHOSPHOLIPASE, PARA-MERCURY-BENZENESULFONIC ACID | | Authors: | Ackerman, S.J, Savage, M.P, Liu, L, Leonidas, D.D, Kwatia, M.A, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2000-11-16 | | Release date: | 2001-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Charcot-Leyden Crystal Protein (Galectin-10) is not a Dual Function Galectin with Lysophospholipase Activity But Binds a Lysophospholipase Inhibitor in a Novel Structural Fashion.
J.Biol.Chem., 277, 2002
|
|
6OUG
 
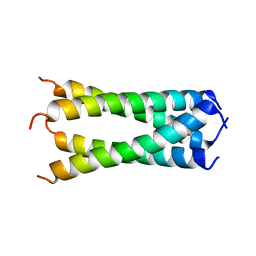 | | Structure of drug-resistant V27A mutant of the influenza M2 proton channel bound to spiroadamantyl amine inhibitor, TM + cytosolic helix construct | | Descriptor: | (1r,1'S,3'S,5'S,7'S)-spiro[cyclohexane-1,2'-tricyclo[3.3.1.1~3,7~]decan]-4-amine, Matrix protein 2 | | Authors: | Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2019-05-04 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | X-ray Crystal Structures of the Influenza M2 Proton Channel Drug-Resistant V27A Mutant Bound to a Spiro-Adamantyl Amine Inhibitor Reveal the Mechanism of Adamantane Resistance.
Biochemistry, 59, 2020
|
|
1KZI
 
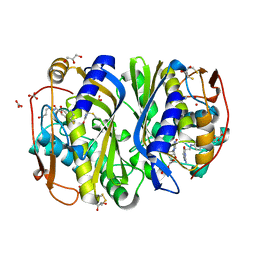 | | Crystal Structure of EcTS/dUMP/THF Complex | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (6S)-5,6,7,8-TETRAHYDROFOLATE, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, ... | | Authors: | Fritz, T.A, Liu, L, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Tryptophan 80 and leucine 143 are critical for the hydride transfer step of thymidylate synthase by controlling active site access.
Biochemistry, 41, 2002
|
|
1KZJ
 
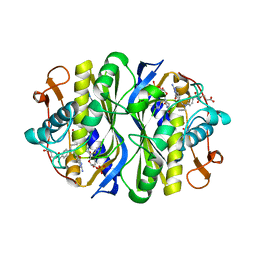 | | Crystal Structure of EcTS W80G/dUMP/CB3717 Complex | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Fritz, T.A, Liu, L, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Tryptophan 80 and leucine 143 are critical for the hydride transfer step of thymidylate synthase by controlling active site access.
Biochemistry, 41, 2002
|
|
1M7Q
 
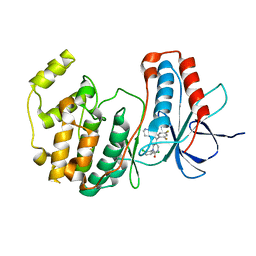 | | Crystal structure of p38 MAP kinase in complex with a dihydroquinazolinone inhibitor | | Descriptor: | 1-(2,6-DICHLOROPHENYL)-5-(2,4-DIFLUOROPHENYL)-7-PIPERAZIN-1-YL-3,4-DIHYDROQUINAZOLIN-2(1H)-ONE, Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Stelmach, J.E, Liu, L, Patel, S.B, Pivnichny, J.V, Scapin, G, Singh, S, Hop, C.E.C.A, Wang, Z, Cameron, P.M, Nichols, E.A, O'Keefe, S.J, O'Neill, E.A, Schmatz, D.M, Schwartz, C.D, Thompson, C.M, Zaller, D.M, Doherty, J.B. | | Deposit date: | 2002-07-22 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of potent, orally bioavailable dihydroquinazolinone inhibitors of p38 MAP kinase.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1VPR
 
 | |
8KAG
 
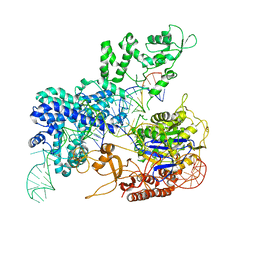 | | Crystal structure of SpyCas9 in complex with sgRNA and target RNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, RNA (5'-R(*CP*CP*AP*CP*UP*UP*CP*AP*AP*UP*UP*AP*GP*AP*AP*CP*AP*CP*GP*GP*AP*CP*C)-3'), RNA (98-MER) | | Authors: | Chen, Y, Chen, J, Liu, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.88 Å) | | Cite: | Trans-nuclease activity of Cas9 activated by DNA or RNA target binding.
Nat.Biotechnol., 2024
|
|
8KAK
 
 | |
8KAJ
 
 | | Crystal structure of SpyCas9-crRNA-tracrRNA complex bound to 16nt target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*CP*AP*AP*TP*AP*CP*CP*TP*TP*TP*TP*AP*TP*CP*CP*AP*TP*AP*AP*AP*TP*TP*CP*G)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*GP*TP*AP*TP*TP*G)-3'), ... | | Authors: | Chen, Y, Chen, J, Liu, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Trans-nuclease activity of Cas9 activated by DNA or RNA target binding.
Nat.Biotechnol., 2024
|
|
8KAI
 
 | |
