7TYU
 
 | | TEAD2 bound to Compound 2 | | Descriptor: | (3R)-1-[(8S)-5-(4-cyclohexylphenyl)-7-oxo-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonyl]pyrrolidine-3-carbonitrile, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Fong, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
7TYP
 
 | | TEAD2 bound to GNE-7883 | | Descriptor: | (8S)-5-(4-cyclohexylphenyl)-3-[3-(fluoromethyl)azetidine-1-carbonyl]-2-(3-methylpyrazin-2-yl)pyrazolo[1,5-a]pyrimidin-7(4H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Fong, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
5DEY
 
 | | Crystal structure of PAK1 in complex with an inhibitor compound G-5555 | | Descriptor: | 8-[(trans-5-amino-1,3-dioxan-2-yl)methyl]-6-[2-chloro-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, Serine/threonine-protein kinase PAK 1 | | Authors: | Oh, A, Tam, C, Wang, W. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2016-06-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of Selective PAK1 Inhibitor G-5555: Improving Properties by Employing an Unorthodox Low-pK a Polar Moiety.
Acs Med.Chem.Lett., 6, 2015
|
|
5DFP
 
 | | Crystal structure of PAK1 in complex with an inhibitor compound FRAX1036 | | Descriptor: | 6-[2-chloro-4-(6-methylpyrazin-2-yl)phenyl]-8-ethyl-2-{[2-(1-methylpiperidin-4-yl)ethyl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, DIMETHYL SULFOXIDE, Serine/threonine-protein kinase PAK 1 | | Authors: | Maksimoska, J, Marmorstein, R, Wang, W. | | Deposit date: | 2015-08-27 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of Selective PAK1 Inhibitor G-5555: Improving Properties by Employing an Unorthodox Low-pK a Polar Moiety.
Acs Med.Chem.Lett., 6, 2015
|
|
2QHB
 
 | |
2CKX
 
 | |
6AK2
 
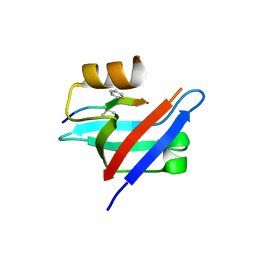 | | Crystal structure of the syntenin PDZ1 domain in complex with the peptide inhibitor KSL-128018 | | Descriptor: | Syntenin-1, peptide inhibitor KSL-128018 | | Authors: | Jin, Z.Y, Park, J.H, Yun, J.H, Haugaard-Kedstrom, L.M, Lee, W.T. | | Deposit date: | 2018-08-29 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | A High-Affinity Peptide Ligand Targeting Syntenin Inhibits Glioblastoma.
J.Med.Chem., 64, 2021
|
|
6UYC
 
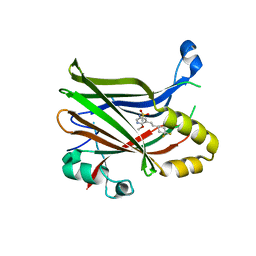 | | Crystal structure of TEAD2 bound to Compound 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-{5-[(E)-2-(4,4-difluorocyclohexyl)ethenyl]-6-methoxypyridin-3-yl}methanesulfonamide, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Holden, J.K, Crawford, J.J, Zbieg, J.R, Cunningham, C.N. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling.
Cell Rep, 31, 2020
|
|
6UYB
 
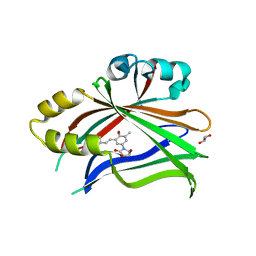 | | Crystal structure of TEAD2 bound to Compound 1 | | Descriptor: | (3R,4R)-1-{3-[(E)-2-(4-chlorophenyl)ethenyl]-4-methoxy-5-methylphenyl}-3,4-dihydroxypyrrolidin-2-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Noland, C.L, Holden, J.K, Crawford, J.J, Zbieg, J.R, Cunningham, C.N. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling.
Cell Rep, 31, 2020
|
|
2XHS
 
 | |
4U81
 
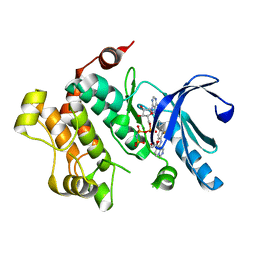 | | MEK1 Kinase bound to small molecule inhibitor G659 | | Descriptor: | 8-[(4-cyclopropyl-2-fluorophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-7-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Robarge, K.D, Ultsch, M.H, Weismann, C. | | Deposit date: | 2014-07-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structure based design of novel 6,5 heterobicyclic mitogen-activated protein kinase kinase (MEK) inhibitors leading to the discovery of imidazo[1,5-a] pyrazine G-479.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4U7Z
 
 | | Mitogen-Activated Protein Kinase Kinase (MEK1) bound to G805 | | Descriptor: | 5-[(4-bromo-2-chlorophenyl)amino]-4-fluoro-N-(2-hydroxyethoxy)-1-methyl-1H-benzimidazole-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Robarge, K.D, Ultsch, M.H, Wiesmann, C. | | Deposit date: | 2014-07-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Structure based design of novel 6,5 heterobicyclic mitogen-activated protein kinase kinase (MEK) inhibitors leading to the discovery of imidazo[1,5-a] pyrazine G-479.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4U80
 
 | | MEK 1 kinase bound to G799 | | Descriptor: | 3-[(4-cyclopropyl-2-fluorophenyl)amino]-N-(2-hydroxyethoxy)furo[3,2-c]pyridine-2-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Ultsch, M.H, Robarge, K.D, Weismann, C. | | Deposit date: | 2014-07-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure based design of novel 6,5 heterobicyclic mitogen-activated protein kinase kinase (MEK) inhibitors leading to the discovery of imidazo[1,5-a] pyrazine G-479.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7CRJ
 
 | | Dark State Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRI
 
 | | 1 ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRL
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 50 ps after light activation | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRY
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (6.49 mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRK
 
 | | 2ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRX
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (2.63mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRT
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.17mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRS
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.90mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7WJ5
 
 | | Cryo-EM structure of human somatostatin receptor 2 complex with its agonist somatostatin delineates the ligand binding specificity | | Descriptor: | Gai1 antibody (scfv16), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Heo, Y.S, Yoon, E.J, Jeon, Y.E, Yun, J.-H, Ishimoto, N, Woo, H, Park, S.Y, Song, J, Lee, W.T. | | Deposit date: | 2022-01-05 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM structure of the human somatostatin receptor 2 complex with its agonist somatostatin delineates the ligand-binding specificity.
Elife, 11, 2022
|
|
1DMQ
 
 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1NE3
 
 | | Solution structure of ribosomal protein S28E from Methanobacterium Thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH0256_1_68; Northeast Structural Genomics Target TT744 | | Descriptor: | 30S ribosomal protein S28E | | Authors: | Wu, B, Pineda-Lucena, A, Yee, A, Cort, J.R, Ramelot, T.A, Kennedy, M, Edwards, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-12-10 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ribosomal protein S28E from Methanobacterium thermoautotrophicum.
Protein Sci., 12, 2003
|
|
1DMM
 
 | | CRYSTAL STRUCTURES OF MUTANT ENZYMES Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
