7BGY
 
 | | Cryo-EM Structure of KdpFABC in E2Pi state with MgF4 | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, MAGNESIUM ION, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BP1
 
 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum in complex with Catechol | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, CATECHOL, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
8P9G
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB390 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-4-[(~{E})-(6-methyl-7-oxidanyl-1~{H}-indol-4-yl)diazenyl]benzamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9I
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB462 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-4-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)benzamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9H
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB437 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-2-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)quinoline-6-carboxamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9K
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB503 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-azanyl-5-phenyl-phenyl)-4-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)benzamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9J
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB500 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-5-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)pyridine-2-carboxamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9F
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB161 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-4-[2-(2-azanyl-5-methyl-4-oxidanyl-phenyl)hydrazinyl]benzamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9L
 
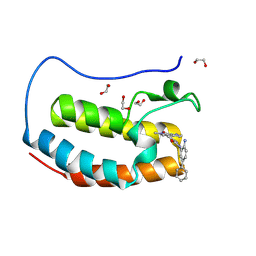 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB512 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-azanyl-5-phenyl-phenyl)-5-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)pyridine-2-carboxamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
7XCK
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 fab (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, S309 light chain, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Xie, Y.F, Liu, S. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCI
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with human ACE2 ectodomain (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCO
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with S309 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Fab heavy chain, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | Deposit date: | 2022-03-24 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
6OOH
 
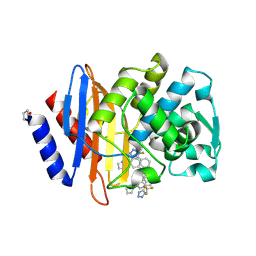 | | CTX-M-27 Beta Lactamase with Compound 14 | | Descriptor: | 3-(1H-pyrazol-1-yl)-N-[3-(1H-tetrazol-5-yl)phenyl]-5-(trifluoromethyl)benzamide, Beta-lactamase | | Authors: | Kemp, M, Chen, Y. | | Deposit date: | 2019-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | An Empirical Study of Amide-Heteroarene pi-Stacking Interactions Using Reversible Inhibitors of a Bacterial Serine Hydrolase.
Org Chem Front, 6, 2019
|
|
8PHI
 
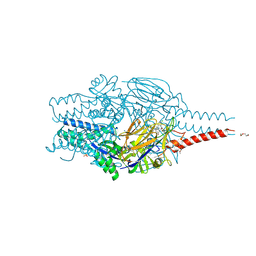 | | Crystal structure of prefusion-stabilized RSV F Variant DS-Cav1 in complex with Lonafarnib | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 4-{2-[4-(3,10-DIBROMO-8-CHLORO-6,11-DIHYDRO-5H-BENZO[5,6]CYCLOHEPTA[1,2-B]PYRIDIN-11-YL)PIPERIDIN-1-YL]-2-OXOETHYL}PIPERIDINE-1-CARBOXAMIDE, CALCIUM ION, ... | | Authors: | Rajak, M, Krey, T. | | Deposit date: | 2023-06-19 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Drug repurposing screen identifies lonafarnib as respiratory syncytial virus fusion protein inhibitor.
Nat Commun, 15, 2024
|
|
8PVQ
 
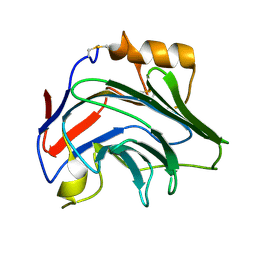 | | Crystal Structure of Human PTX3 C-terminal domain | | Descriptor: | Pentraxin-related protein PTX3 | | Authors: | Levy, C.W. | | Deposit date: | 2023-07-18 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Pentraxin 3 mediated crosslinking of heavy chain hyaluronan complexes: new insights from structural and biophysical studies
To Be Published
|
|
6OOJ
 
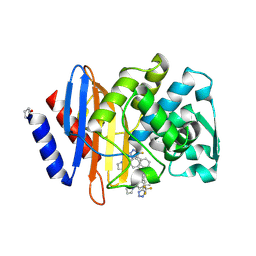 | | CTX-M-14 Beta Lactamase with Compound 14 | | Descriptor: | 3-(1H-pyrazol-1-yl)-N-[3-(1H-tetrazol-5-yl)phenyl]-5-(trifluoromethyl)benzamide, Beta-lactamase | | Authors: | Kemp, M, Chen, Y. | | Deposit date: | 2019-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Empirical Study of Amide-Heteroarene pi-Stacking Interactions Using Reversible Inhibitors of a Bacterial Serine Hydrolase.
Org Chem Front, 6, 2019
|
|
8PEF
 
 | |
6OOK
 
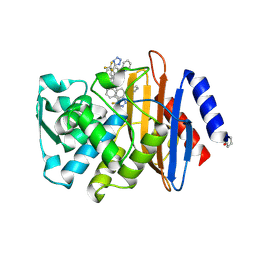 | | CTX-M-14 Beta Lactamase with Compound 3 | | Descriptor: | 3-(pyridin-2-yl)-N-[3-(1H-tetrazol-5-yl)phenyl]-5-(trifluoromethyl)benzamide, Beta-lactamase, PHOSPHATE ION | | Authors: | Kemp, M, Chen, Y. | | Deposit date: | 2019-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Empirical Study of Amide-Heteroarene pi-Stacking Interactions Using Reversible Inhibitors of a Bacterial Serine Hydrolase.
Org Chem Front, 6, 2019
|
|
5T00
 
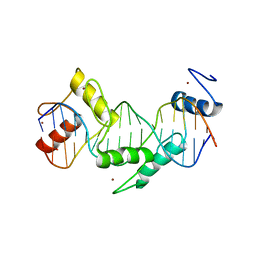 | | Human CTCF ZnF3-7 and methylated DNA complex | | Descriptor: | DNA (5'-GCCAGCAGGGGG(5CM)GCTA-3'), DNA (5'-TAG(5CM)GCCCCCTGCTGGC-3'), Transcriptional repressor CTCF, ... | | Authors: | Hashimoto, H, Wang, D, Cheng, X. | | Deposit date: | 2016-08-15 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
5T01
 
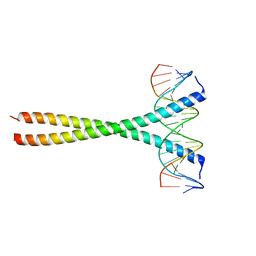 | | Human c-Jun DNA binding domain homodimer in complex with methylated DNA | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*GP*AP*(5CM)P*GP*AP*GP*TP*CP*AP*TP*AP*GP*GP*AP*G)-3'), DNA (5'-D(P*CP*TP*CP*CP*TP*AP*TP*GP*AP*CP*TP*CP*GP*TP*CP*CP*AP*T)-3'), Transcription factor AP-1 | | Authors: | Hong, S, Horton, J.R, Cheng, X. | | Deposit date: | 2016-08-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
8QEX
 
 | |
7E3J
 
 | |
7XLB
 
 | |
6OOE
 
 | | CTX-M-27 Beta Lactamase with Compound 20 | | Descriptor: | 3-(1H-tetrazol-5-yl)-N-[3-(1H-tetrazol-5-yl)phenyl]-5-(trifluoromethyl)benzamide, Beta-lactamase | | Authors: | Kemp, M, Chen, Y. | | Deposit date: | 2019-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | An Empirical Study of Amide-Heteroarene pi-Stacking Interactions Using Reversible Inhibitors of a Bacterial Serine Hydrolase.
Org Chem Front, 6, 2019
|
|
6OOF
 
 | | CTX-M-14 Beta Lactamase with Compound 20 | | Descriptor: | 3-(1H-tetrazol-5-yl)-N-[3-(1H-tetrazol-5-yl)phenyl]-5-(trifluoromethyl)benzamide, Beta-lactamase | | Authors: | Kemp, M, Chen, Y. | | Deposit date: | 2019-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.236 Å) | | Cite: | An Empirical Study of Amide-Heteroarene pi-Stacking Interactions Using Reversible Inhibitors of a Bacterial Serine Hydrolase.
Org Chem Front, 6, 2019
|
|
