7F1O
 
 | |
7F1Z
 
 | |
7CN0
 
 | | Cryo-EM structure of K+-bound hERG channel | | Descriptor: | POTASSIUM ION, potassium channel 1 | | Authors: | Asai, T, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Senda, T, Murata, T. | | Deposit date: | 2020-07-29 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Structure of K + -Bound hERG Channel Complexed with the Blocker Astemizole.
Structure, 29, 2021
|
|
2OJU
 
 | | X-ray structure of complex of human cyclophilin J with cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE-LIKE 3 | | Authors: | Xia, Z, Huang, L. | | Deposit date: | 2007-01-14 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting Cyclophilin J, a Novel Peptidyl-Prolyl Isomerase, Can Induce Cellular G1/S Arrest and Repress the Growth of Hepatocellular Carcinoma
To be Published
|
|
2PVP
 
 | |
4D3O
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6-(3-(2-(1H-Pyrrolo(2,3-b)pyridin-6-yl)ethyl)-5-(aminomethyl) phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 6-(2-{3-(aminomethyl)-5-[2-(1H-pyrrolo[2,3-b]pyridin-6-yl)ethyl]phenyl}ethyl)-4-methylpyridin-2-amine, CHLORIDE ION, GLYCEROL, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-10-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of Bacterial Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 58, 2015
|
|
4LH5
 
 | | Dual inhibition of HIV-1 replication by Integrase-LEDGF allosteric inhibitors is predominant at post-integration stage during virus production rather than at integration | | Descriptor: | (2S)-tert-butoxy[4-(3,4-dihydro-2H-chromen-6-yl)-2-methylquinolin-3-yl]ethanoic acid, Integrase, MAGNESIUM ION | | Authors: | Ruff, M, Levy, N, Eiler, S. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Dual inhibition of HIV-1 replication by integrase-LEDGF allosteric inhibitors is predominant at the post-integration stage.
Retrovirology, 10, 2013
|
|
4LH4
 
 | |
3O64
 
 | | Crystal structure of catalytic domain of TACE with 2-(2-Aminothiazol-4-yl)pyrrolidine-Based Tartrate Diamides | | Descriptor: | (2R,3R)-2,3-dihydroxy-4-{(2R)-2-[2-(methylamino)-5-(methylsulfonyl)-1,3-thiazol-4-yl]pyrrolidin-1-yl}-4-oxo-N-{(1R)-1-[4-(1H-pyrazol-1-yl)phenyl]ethyl}butanamide, CALCIUM ION, ISOPROPYL ALCOHOL, ... | | Authors: | Orth, P. | | Deposit date: | 2010-07-28 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 2-(2-Aminothiazol-4-yl)pyrrolidine-based tartrate diamides as potent, selective and orally bioavailable TACE inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3O3T
 
 | | Crystal Structure Analysis of M32A mutant of human CLIC1 | | Descriptor: | Chloride intracellular channel protein 1 | | Authors: | Fanucchi, S, Achilonu, I.A, Adamson, R.J, Fernandes, M.A, Stoychev, S, Dirr, H.W. | | Deposit date: | 2010-07-26 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure Analysis of M32A mutant of human CLIC1
To be Published
|
|
8DVH
 
 | | Crystal structure of ATP-dependent Lon protease from Bacillus subtillis (BsLonBA) | | Descriptor: | Lon protease 2, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, SODIUM ION | | Authors: | Sekula, B, Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unique Structural Fold of LonBA Protease from Bacillus subtilis, a Member of a Newly Identified Subfamily of Lon Proteases.
Int J Mol Sci, 23, 2022
|
|
6KX2
 
 | | Crystal structure of GDP bound RhoA protein | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
6KX3
 
 | | Crystal structure of RhoA protein with covalent inhibitor DC-Rhoin | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, prop-2-enyl (3R)-1,1-bis(oxidanylidene)-2,3-dihydro-1-benzothiophene-3-carboxylate | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
4YJN
 
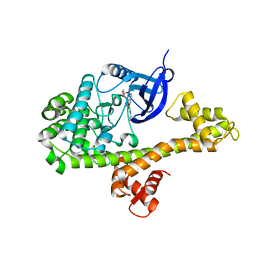 | |
3PJA
 
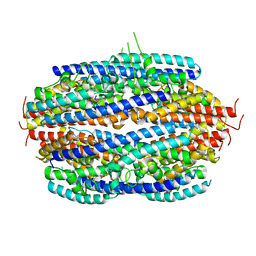 | | Crystal structure of human C3PO complex | | Descriptor: | Translin, Translin-associated protein X | | Authors: | Huang, N, Zhang, H. | | Deposit date: | 2010-11-09 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of C3PO and mechanism of human RISC activation.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3CXC
 
 | | The structure of an enhanced oxazolidinone inhibitor bound to the 50S ribosomal subunit of H. marismortui | | Descriptor: | (3Z)-N-[(4E)-5-(4-{(5S)-5-[(acetylamino)methyl]-2-oxo-1,3-oxazolidin-3-yl}-2-fluorophenyl)pent-4-en-1-yl]-3-(4-methyl-2,6-dioxo-1,6-dihydropyrimidin-5(2H)-ylidene)propanamide, 23S RIBOSOMAL RNA, 5'-R(*CP*CP*A)-3', ... | | Authors: | Ippolito, J.A, Wang, D, Kanyo, Z.F, Duffy, E.M. | | Deposit date: | 2008-04-24 | | Release date: | 2009-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design at the atomic level: design of biaryloxazolidinones as potent orally active antibiotics.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7ENQ
 
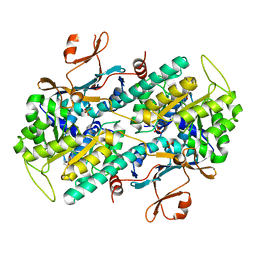 | | Crystal structure of human NAMPT in complex with compound NAT | | Descriptor: | 2-(2-~{tert}-butylphenoxy)-~{N}-(4-hydroxyphenyl)ethanamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Wang, G, Wu, C, Liu, M, Yao, H, Li, C, Wang, L, Tang, Y. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.204966 Å) | | Cite: | Discovery of small-molecule activators of nicotinamide phosphoribosyltransferase (NAMPT) and their preclinical neuroprotective activity.
Cell Res., 32, 2022
|
|
4P5E
 
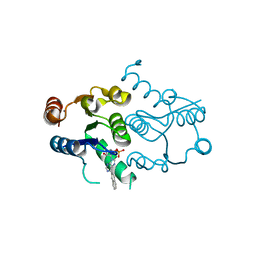 | | CRYSTAL STRUCTURE OF HUMAN DNPH1 (RCL) WITH 6-NAPHTHYL-PURINE-RIBOSIDE-MONOPHOSPHATE | | Descriptor: | 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, 6-(naphthalen-2-yl)-9-(5-O-phosphono-beta-D-ribofuranosyl)-9H-purine, CALCIUM ION | | Authors: | Padilla, A, Labesse, G, Kaminski, P.A. | | Deposit date: | 2014-03-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | 6-(Hetero)Arylpurine nucleotides as inhibitors of the oncogenic target DNPH1: Synthesis, structural studies and cytotoxic activities.
Eur.J.Med.Chem., 85C, 2014
|
|
5H8T
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with all-trans-retinol | | Descriptor: | RETINOL, Retinol-binding protein 1 | | Authors: | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | Deposit date: | 2015-12-23 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
5HBS
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with all-trans-retinol at 0.89 angstrom. | | Descriptor: | RETINOL, Retinol-binding protein 1 | | Authors: | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | Deposit date: | 2016-01-02 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
5HA1
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with retinylamine | | Descriptor: | (2~{E},4~{E},6~{E},8~{E})-3,7-dimethyl-9-(2,6,6-trimethylcyclohexen-1-yl)nona-2,4,6,8-tetraen-1-amine, Retinol-binding protein 1 | | Authors: | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | Deposit date: | 2015-12-29 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
6MBW
 
 | | Structure of Transcription Factor | | Descriptor: | Signal transducer and activator of transcription 5B | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2018-08-30 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural and functional consequences of the STAT5BN642H driver mutation.
Nat Commun, 10, 2019
|
|
6MBZ
 
 | | Structure of Transcription Factor | | Descriptor: | Signal transducer and activator of transcription 5B | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2018-08-30 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural and functional consequences of the STAT5BN642Hdriver mutation.
Nat Commun, 10, 2019
|
|
4P5D
 
 | | CRYSTAL STRUCTURE OF RAT DNPH1 (RCL) WITH 6-NAPHTHYL-PURINE-RIBOSIDE-MONOPHOSPHATE | | Descriptor: | 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, 6-(naphthalen-2-yl)-9-(5-O-phosphono-beta-D-ribofuranosyl)-9H-purine, SULFATE ION | | Authors: | Padilla, A, Labesse, G, Kaminski, P.A. | | Deposit date: | 2014-03-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | 6-(Hetero)Arylpurine nucleotides as inhibitors of the oncogenic target DNPH1: Synthesis, structural studies and cytotoxic activities.
Eur.J.Med.Chem., 85C, 2014
|
|
5AO1
 
 | |
