8ZMY
 
 | | F0502B-bound WT polymorph 5a alpha-synuclein fibril | | Descriptor: | 2-bromanyl-4-[(~{E})-2-[6-[2-(2-fluoranylethoxy)ethyl-methyl-amino]-5-methyl-1,3-benzothiazol-2-yl]ethenyl]phenol, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2024-05-24 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6LFO
 
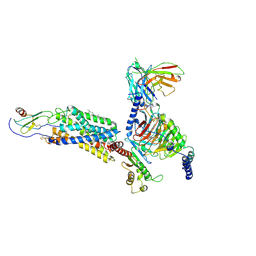 | | Cryo-EM structure of a class A GPCR monomer | | Descriptor: | C-X-C chemokine receptor type 2, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Z.J, Hua, T, Liu, K.W, Wu, L.J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of CXC chemokine receptor 2 activation and signalling.
Nature, 585, 2020
|
|
6LFM
 
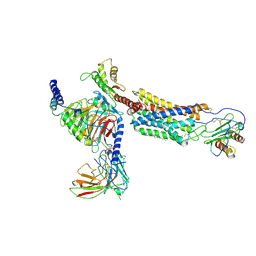 | | Cryo-EM structure of a class A GPCR | | Descriptor: | C-X-C chemokine receptor type 2, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Z.J, Hua, T, Liu, K.W, Wu, L.J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of CXC chemokine receptor 2 activation and signalling.
Nature, 585, 2020
|
|
6LFL
 
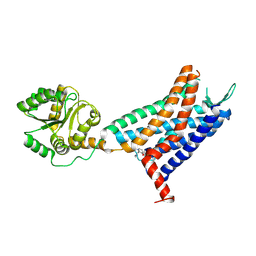 | | Crystal structure of a class A GPCR | | Descriptor: | 4-[[3,4-bis(oxidanylidene)-2-[[(1~{R})-1-(4-propan-2-ylfuran-2-yl)propyl]amino]cyclobuten-1-yl]amino]-~{N},~{N}-dimethyl-3-oxidanyl-pyridine-2-carboxamide, C-X-C chemokine receptor type 2,GlgA glycogen synthase,C-X-C chemokine receptor type 2 | | Authors: | Liu, Z.J, Hua, T, Liu, K.W, Wu, L.J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of CXC chemokine receptor 2 activation and signalling.
Nature, 585, 2020
|
|
3PMT
 
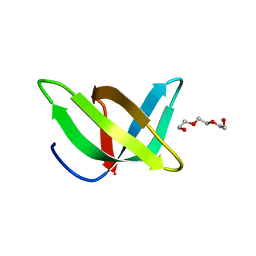 | | Crystal structure of the Tudor domain of human Tudor domain-containing protein 3 | | Descriptor: | TETRAETHYLENE GLYCOL, Tudor domain-containing protein 3 | | Authors: | Lam, R, Bian, C.B, Guo, Y.H, Xu, C, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of TDRD3 and Methyl-Arginine Binding Characterization of TDRD3, SMN and SPF30.
Plos One, 7, 2012
|
|
3S6W
 
 | | Crystal structure of Tudor domain of human TDRD3 | | Descriptor: | ISOPROPYL ALCOHOL, Tudor domain-containing protein 3 | | Authors: | Liu, H.P, Xu, R.M. | | Deposit date: | 2011-05-26 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of TDRD3 and methyl-arginine binding characterization of TDRD3, SMN and SPF30
Plos One, 7, 2012
|
|
8X7P
 
 | | CCA-bound E46K alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein, copper;trisodium;18-(2-carboxylatoethyl)-20-(carboxylatomethyl)-12-ethenyl-7-ethyl-3,8,13,17-tetramethyl-17,18-dihydroporphyrin-21,23-diide-2-carboxylate | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7M
 
 | | CR-bound E46K alpha-synuclein fibrils | | Descriptor: | 4-azanyl-3-[(~{E})-[4-[4-[(~{E})-(1-azanyl-4-sulfo-naphthalen-2-yl)diazenyl]phenyl]phenyl]diazenyl]naphthalene-1-sulfonic acid, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7O
 
 | | PiB-bound E46K mutanted alpha-synuclein fibrils | | Descriptor: | 2-[4-(methylamino)phenyl]-1,3-benzothiazol-6-ol, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7R
 
 | | C05-03-bound E46K alpha-synuclein fibrils | | Descriptor: | 2-[(~{E})-4-[6-(methylamino)pyridin-3-yl]but-1-en-3-ynyl]-1,3-benzothiazol-6-ol, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7L
 
 | | EB-bound E46K alpha-synuclein fibrils | | Descriptor: | 4-azanyl-6-[[4-[4-[(~{E})-(8-azanyl-1-oxidanyl-5,7-disulfo-naphthalen-2-yl)diazenyl]-3-methyl-phenyl]-2-methyl-phenyl]diazenyl]-5-oxidanyl-naphthalene-1,3-disulfonic acid, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7Q
 
 | | pFTAA-bound E46K alpha-synuclein fibrils | | Descriptor: | 3''',4'-bis(carboxymethyl)-2,2':5',2'':5'',2''':5''',2''''-quinquethiophene-5,5''''-dicarboxylic acid, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6C1U
 
 | | MBD2 in complex with a deoxy-oligonucleotide | | Descriptor: | 12-mer DNA, Methyl-CpG-binding domain protein 2, UNKNOWN ATOM OR ION | | Authors: | Lei, M, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-05 | | Release date: | 2018-02-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the ability of MBD domains to bind methyl-CG and TG sites in DNA.
J. Biol. Chem., 293, 2018
|
|
6CNP
 
 | |
6C1T
 
 | | MBD2 in complex with a partially methylated DNA | | Descriptor: | 12-mer DNA, GLYCEROL, Methyl-CpG-binding domain protein 2, ... | | Authors: | Lei, M, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-05 | | Release date: | 2018-02-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the ability of MBD domains to bind methyl-CG and TG sites in DNA.
J. Biol. Chem., 293, 2018
|
|
6C1V
 
 | | MBD2 in complex with double-stranded DNA | | Descriptor: | 12-mer DNA, Methyl-CpG-binding domain protein 2, UNKNOWN ATOM OR ION | | Authors: | Lei, M, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-05 | | Release date: | 2018-02-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the ability of MBD domains to bind methyl-CG and TG sites in DNA.
J. Biol. Chem., 293, 2018
|
|
7C8J
 
 | | Structural basis for cross-species recognition of COVID-19 virus spike receptor binding domain to bat ACE2 | | Descriptor: | Angiotensin-converting enzyme, SARS-CoV-2 Receptor binding domain, ZINC ION | | Authors: | Liu, K.F, Wang, J, Tan, S.G, Niu, S, Wu, L.L, Zhang, Y.F, Pan, X.Q, Meng, Y.M, Chen, Q, Wang, Q.H, Wang, H.W, Qi, J.X, Gao, G.F. | | Deposit date: | 2020-06-01 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Cross-species recognition of SARS-CoV-2 to bat ACE2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7C8K
 
 | | Structural basis for cross-species recognition of COVID-19 virus spike receptor binding domain to bat ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Liu, K.F, Wang, J, Tan, S.G, Niu, S, Wu, L.L, Zhang, Y.F, Pan, X.Q, Meng, Y.M, Chen, Q, Wang, Q.H, Wang, H.W, Qi, J.X, Gao, G.F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cross-species recognition of SARS-CoV-2 to bat ACE2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CU5
 
 | | N-Glycosylation of PD-1 and glycosylation dependent binding of PD-1 specific monoclonal antibody camrelizumab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Programmed cell death protein 1, ... | | Authors: | Liu, K.F, Tan, S.G, Jin, W.J, Guan, J.W, Wang, W.L, Sun, H, Qi, J.X, Yan, J.H, Chai, Y, Wang, Z.F, Chu, X.D, Gao, G.F. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | N-glycosylation of PD-1 promotes binding of camrelizumab.
Embo Rep., 21, 2020
|
|
7DRV
 
 | | Structural basis of SARS-CoV-2-closely-related bat coronavirus RaTG13 to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liu, K.F, Pan, X.Q, Li, L.J, Feng, Y, Meng, Y.M, Zhang, Y.F, Wu, L.L, Chen, Q, Zheng, A.Q, Song, C.L, Jia, Y.F, Niu, S, Qiao, C.P, Zhao, X, Ma, D.L, Ma, X.P, Tan, S.G, Qi, J.X, Gao, G.F, Wang, Q.H. | | Deposit date: | 2020-12-29 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species.
Cell, 184, 2021
|
|
5IZU
 
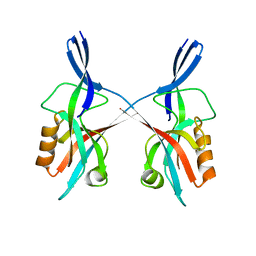 | |
5J39
 
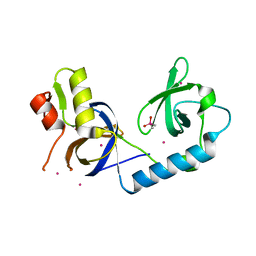 | | Crystal Structure of the extended TUDOR domain from TDRD2 | | Descriptor: | CACODYLATE ION, Tudor and KH domain-containing protein, UNKNOWN ATOM OR ION | | Authors: | Zhang, H, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-30 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for arginine methylation-independent recognition of PIWIL1 by TDRD2.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7VUG
 
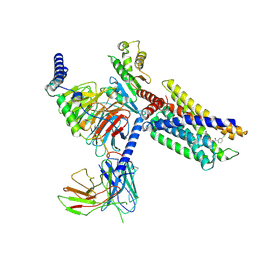 | | Cryo-EM structure of a class A orphan GPCR in complex with Gi | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Chimera of Endo-1,4-beta-xylanase and Probable G-protein coupled receptor 139, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUH
 
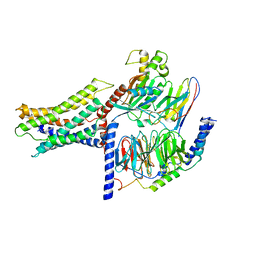 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUJ
 
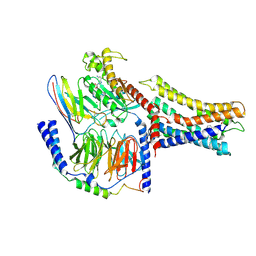 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
