4LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
6H1V
 
 | |
6QIB
 
 | |
2NOY
 
 | | Crystal structure of transthyretin mutant I84S at PH 7.5 | | Descriptor: | Transthyretin | | Authors: | Pasquato, N, Berni, R, Folli, C, Alfieri, B, Cendron, L, Zanotti, G. | | Deposit date: | 2006-10-26 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acidic pH-induced conformational changes in amyloidogenic mutant transthyretin
J.Mol.Biol., 366, 2007
|
|
2G3X
 
 | |
2G4G
 
 | |
2G4E
 
 | |
2G3Z
 
 | |
3BT0
 
 | | Crystal structure of transthyretin variant V20S | | Descriptor: | Transthyretin | | Authors: | Zanotti, G, Folli, C, Cendron, L, Gliubich, F, Negro, A, Berni, R. | | Deposit date: | 2007-12-27 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural and mutational analyses of protein-protein interactions between transthyretin and retinol-binding protein.
Febs J., 275, 2008
|
|
3DJS
 
 | |
3DJZ
 
 | |
3CXF
 
 | | Crystal structure of transthyretin variant Y114H | | Descriptor: | Transthyretin | | Authors: | Cendron, L, Zanotti, G, Folli, C, Alfieri, B, Pasquato, N, Berni, R. | | Deposit date: | 2008-04-24 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational analyses of protein-protein interactions between transthyretin and retinol-binding protein.
Febs J., 275, 2008
|
|
3DJR
 
 | |
3DJT
 
 | |
3DK0
 
 | |
3DK2
 
 | |
3DO4
 
 | |
3T5U
 
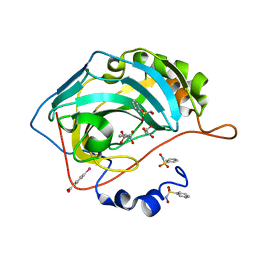 | | Crystal structure of the human carbonic anhydrase II in complex with N-hydroxy benzenesulfonamide | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, MERCURIBENZOIC ACID, ... | | Authors: | Di Fiore, A, Maresca, A, Alterio, V, Supuran, C.T, De Simone, G. | | Deposit date: | 2011-07-28 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Carbonic anhydrase inhibitors: X-ray crystallographic studies for the binding of N-substituted benzenesulfonamides to human isoform II.
Chem.Commun.(Camb.), 47, 2011
|
|
3T5Z
 
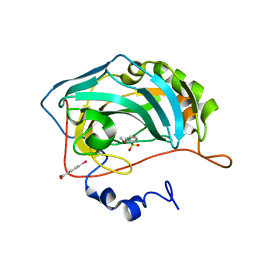 | | Crystal structure of the human carbonic anhydrase II in complex with N-methoxy-benzenesulfonamide | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, N-methoxybenzenesulfonamide, ... | | Authors: | Di Fiore, A, Maresca, A, Alterio, V, Supuran, C.T, De Simone, G. | | Deposit date: | 2011-07-28 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Carbonic anhydrase inhibitors: X-ray crystallographic studies for the binding of N-substituted benzenesulfonamides to human isoform II.
Chem.Commun.(Camb.), 47, 2011
|
|
1OO2
 
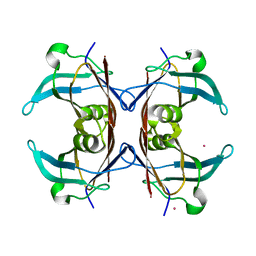 | | Crystal structure of transthyretin from Sparus aurata | | Descriptor: | CADMIUM ION, transthyretin | | Authors: | Pasquato, N, Ramazzina, I, Folli, C, Battistutta, R, Berni, R, Zanotti, G. | | Deposit date: | 2003-03-03 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Distinctive binding and structural properties of piscine transthyretin.
Febs Lett., 555, 2003
|
|
1CVK
 
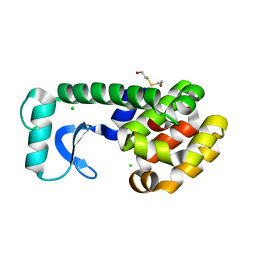 | | T4 LYSOZYME MUTANT L118A | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-23 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
2HD6
 
 | | Crystal structure of the human carbonic anhydrase II in complex with a hypoxia-activatable sulfonamide. | | Descriptor: | CHLORIDE ION, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | De Simone, G, Vitale, R.M, Di Fiore, A, Pedone, C. | | Deposit date: | 2006-06-20 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Carbonic anhydrase inhibitors: Hypoxia-activatable sulfonamides incorporating disulfide bonds that target the tumor-associated isoform IX.
J.Med.Chem., 49, 2006
|
|
2HNC
 
 | | Crystal structure of the human carbonic anhydrase II in complex with the 5-amino-1,3,4-thiadiazole-2-sulfonamide inhibitor. | | Descriptor: | 5-AMINO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Menchise, V, Di Fiore, A, De Simone, G. | | Deposit date: | 2006-07-12 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Carbonic anhydrase inhibitors: X-ray crystallographic studies for the binding of 5-amino-1,3,4-thiadiazole-2-sulfonamide and 5-(4-amino-3-chloro-5-fluorophenylsulfonamido)-1,3,4-thiadiazole-2-sulfonamide to human isoform II.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2HL4
 
 | | Crystal structure analysis of human carbonic anhydrase II in complex with a benzenesulfonamide derivative | | Descriptor: | CHLORIDE ION, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Di Fiore, A, Supuran, C.T, Winum, J.-Y, Montero, J.-L, Pedone, C, Scozzafava, A, De Simone, G. | | Deposit date: | 2006-07-06 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Carbonic anhydrase inhibitors: binding of an antiglaucoma glycosyl-sulfanilamide derivative to human isoform II and its consequences for the drug design of enzyme inhibitors incorporating sugar moieties
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2HOC
 
 | | Crystal structure of the human carbonic anhydrase II in complex with the 5-(4-amino-3-chloro-5-fluorophenylsulfonamido)-1,3,4-thiadiazole-2-sulfonamide inhibitor | | Descriptor: | 5-{[(4-AMINO-3-CHLORO-5-FLUOROPHENYL)SULFONYL]AMINO}-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Menchise, V, Di Fiore, A, De Simone, G. | | Deposit date: | 2006-07-14 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carbonic anhydrase inhibitors: X-ray crystallographic studies for the binding of 5-amino-1,3,4-thiadiazole-2-sulfonamide and 5-(4-amino-3-chloro-5-fluorophenylsulfonamido)-1,3,4-thiadiazole-2-sulfonamide to human isoform II.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
