6CD4
 
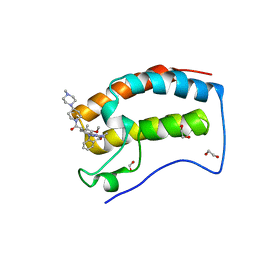 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JWG046 | | Descriptor: | 1,2-ETHANEDIOL, 2-({2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-11-(propan-2-yl)-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2018-02-08 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
6CD5
 
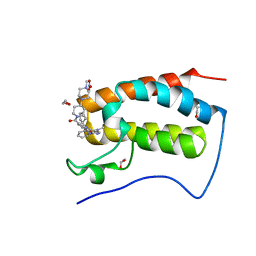 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor XMD17-26 | | Descriptor: | 1,2-ETHANEDIOL, 11-cyclopentyl-2-[[2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl-phenyl]amino]-5-methyl-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4, ... | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2018-02-08 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
6CJ2
 
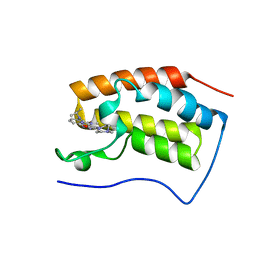 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JWG056 | | Descriptor: | 2-{[3-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}-5-methyl-11-(propan-2-yl)-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
Acs Chem.Biol., 13, 2018
|
|
6CJ1
 
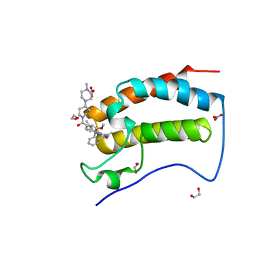 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JWG071 | | Descriptor: | 1,2-ETHANEDIOL, 11-[(2R)-butan-2-yl]-2-({2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, 11-[(2S)-butan-2-yl]-2-({2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, ... | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2018-02-25 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
7FC0
 
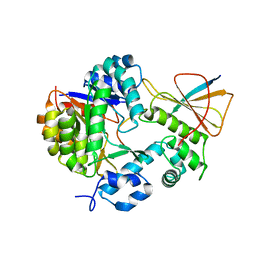 | | Reconstitution of MbnABC complex from Rugamonas rubra ATCC-43154 (GroupIII) | | Descriptor: | FE (III) ION, Methanobactin biosynthesis cassette protein MbnB, Methanobactin biosynthesis cassette protein MbnC, ... | | Authors: | Chao, D, Zhaolin, L, Shoujie, L, Li, Z, Dan, Z, Ying, J, Wei, C. | | Deposit date: | 2021-07-13 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.643 Å) | | Cite: | Crystal structure and catalytic mechanism of the MbnBC holoenzyme required for methanobactin biosynthesis.
Cell Res., 32, 2022
|
|
8HRD
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant in complex with IMCAS74 Fab and W14 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS74 Fab heavy chain, IMCAS74 Fab light chain, ... | | Authors: | Zhao, R.C, Wu, L.L, Han, P. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8XJV
 
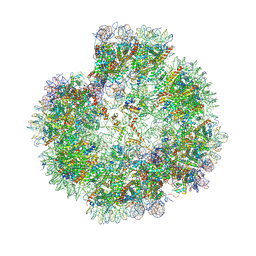 | | Structural basis for the linker histone H5-nucleosome binding and chromatin compaction | | Descriptor: | DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, W.Y, Song, F, Zhu, P. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for linker histone H5-nucleosome binding and chromatin fiber compaction.
Cell Res., 34, 2024
|
|
7DZ9
 
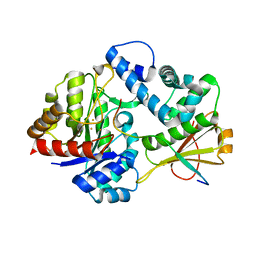 | | MbnABC complex | | Descriptor: | FE (III) ION, MbnA, MbnB, ... | | Authors: | Chao, D, Dan, Z, Yijun, G, Wei, C. | | Deposit date: | 2021-01-25 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and catalytic mechanism of the MbnBC holoenzyme required for methanobactin biosynthesis.
Cell Res., 32, 2022
|
|
7CWQ
 
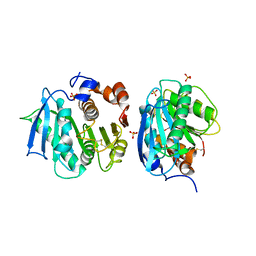 | | Crystal structure of a novel cutinase from Burkhoderiales bacterium RIFCSPLOWO2_02_FULL_57_36 | | Descriptor: | DLH domain-containing protein, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2020-08-30 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | General features to enhance enzymatic activity of poly(ethylene terephthalate) hydrolysis.
Nat Catal, 4, 2021
|
|
7CY0
 
 | | Crystal structure of S185H mutant PET hydrolase from Ideonella sakaiensis | | Descriptor: | ACETIC ACID, Poly(ethylene terephthalate) hydrolase | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | General features to enhance enzymatic activity of poly(ethylene terephthalate) hydrolysis.
Nat Catal, 4, 2021
|
|
7Y3N
 
 | | Crystal structure of SARS-CoV receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, ... | | Authors: | Rao, X, Chai, Y, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7Y3O
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, Light chain of BIOLS56, ... | | Authors: | Rao, X, Gao, F, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7W44
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_RIP | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Leaf-branch compost cutinase | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7VVC
 
 | | Crystal structure of inactive mutant of leaf-branch compost cutinase variant | | Descriptor: | ACETATE ION, ACETIC ACID, CALCIUM ION, ... | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W45
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KIP | | Descriptor: | CALCIUM ION, Leaf-branch compost cutinase, SODIUM ION | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W1N
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KRP | | Descriptor: | 1,2-ETHANEDIOL, BICINE, Leaf-branch compost cutinase | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
8W6G
 
 | | NaDC1 with inhibitor ACA | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-[[(~{E})-3-(4-pentylphenyl)prop-2-enoyl]amino]benzoic acid, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8W6T
 
 | | NaS1 in IN/OUT state | | Descriptor: | Solute carrier family 13 member 1 | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-29 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8W6H
 
 | | NaS1 with sulfate - IN/IN state | | Descriptor: | CHOLESTEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8XXW
 
 | | Fab M2-7 complexed with SARS-Cov2 RBD and human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, M2-7-Heavy chain, M2-7-Light chain, ... | | Authors: | Liu, C, Xie, Y. | | Deposit date: | 2024-01-19 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Mosaic RBD nanoparticle elicits immunodominant antibody responses across sarbecoviruses.
Cell Rep, 43, 2024
|
|
8W6D
 
 | | CryoEM structure of NaDC1 in apo state | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8W6N
 
 | | NaS1 with sulfate in IN/OUT state | | Descriptor: | SODIUM ION, SULFATE ION, Solute carrier family 13 member 1 | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-29 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8W6O
 
 | | NaS1 in IN/IN state | | Descriptor: | CHOLESTEROL, SODIUM ION, Solute carrier family 13 member 1 | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-29 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8HPU
 
 | |
8HQ7
 
 | |
