8I6M
 
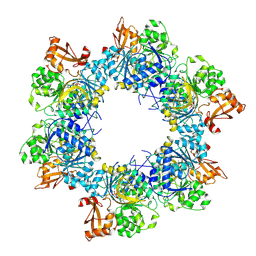 | | Acyl-ACP synthetase structure bound to AMP-C18:1 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, MAGNESIUM ION, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-28 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Acyl-ACP synthetase structure bound to AMP-C18:1
To Be Published
|
|
8I49
 
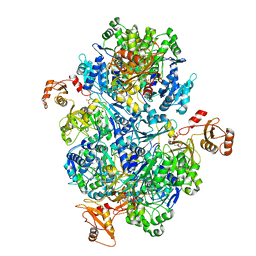 | | Acyl-ACP synthetase structure bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Acyl-ACP synthetase structure bound to ATP.
To Be Published
|
|
8HZX
 
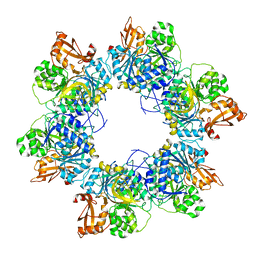 | | Acyl-ACP synthetase structure-2 | | Descriptor: | Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Acyl-ACP synthetase structure-2
To Be Published
|
|
8I8D
 
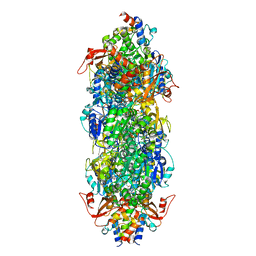 | | Acyl-ACP synthetase structure bound to MC7-ACP | | Descriptor: | 7-methoxy-7-oxidanylidene-heptanoic acid, ADENOSINE MONOPHOSPHATE, Acyl carrier protein, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Acyl-ACP synthetase structure bound to MC7-ACP
To Be Published
|
|
8I8E
 
 | | Acyl-ACP synthetase structure bound to C18:1-ACP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE MONOPHOSPHATE, Acyl carrier protein, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-02-04 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Acyl-ACP synthetase structure bound to C18:1-ACP
To Be Published
|
|
3C97
 
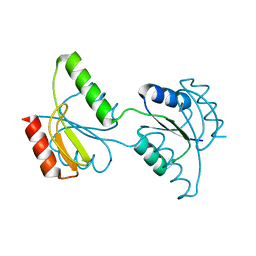 | | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae | | Descriptor: | Signal transduction histidine kinase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-15 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae.
To be Published
|
|
3EEQ
 
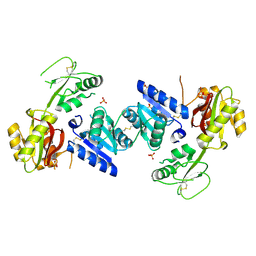 | | Crystal structure of a putative cobalamin biosynthesis protein G homolog from Sulfolobus solfataricus | | Descriptor: | SULFATE ION, putative Cobalamin biosynthesis protein G homolog | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-05 | | Release date: | 2008-09-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative cobalamin biosynthesis protein G homolog from Sulfolobus solfataricus
To be Published
|
|
3EDM
 
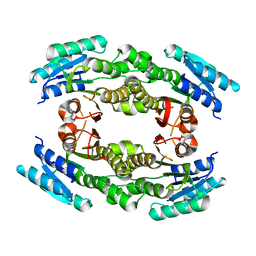 | | Crystal structure of a short chain dehydrogenase from Agrobacterium tumefaciens | | Descriptor: | Short chain dehydrogenase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-03 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a short chain dehydrogenase from Agrobacterium tumefaciens
To be Published
|
|
3CLW
 
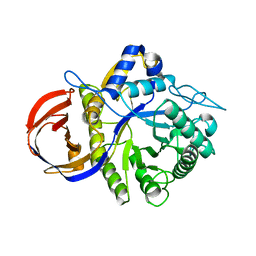 | | Crystal structure of conserved exported protein from Bacteroides fragilis | | Descriptor: | Conserved exported protein | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Chang, S, Ozyurt, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Crystal structure of conserved exported protein from Bacteroides fragilis.
To be Published
|
|
3N3D
 
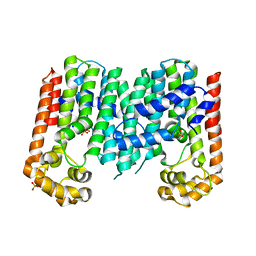 | | Crystal structure of geranylgeranyl pyrophosphate synthase from lactobacillus brevis atcc 367 | | Descriptor: | Geranylgeranyl pyrophosphate synthase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-19 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase from Lactobacillus Brevis
To be Published
|
|
3CYM
 
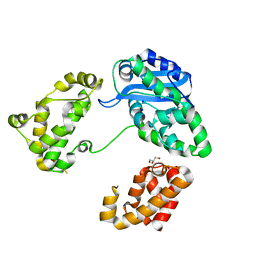 | | Crystal structure of protein BAD_0989 from Bifidobacterium adolescentis | | Descriptor: | GLYCEROL, SODIUM ION, Uncharacterized protein BAD_0989 | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Chang, S, Bain, K, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of protein BAD_0989 from Bifidobacterium adolescentis.
To be Published
|
|
3E61
 
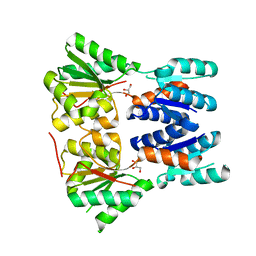 | | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus | | Descriptor: | GLYCEROL, Putative transcriptional repressor of ribose operon | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-14 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus
To be Published
|
|
3G12
 
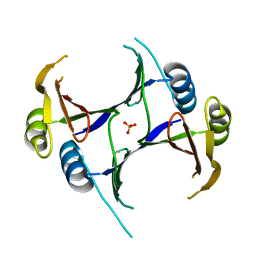 | | Crystal structure of a putative lactoylglutathione lyase from Bdellovibrio bacteriovorus | | Descriptor: | Putative lactoylglutathione lyase, SULFATE ION | | Authors: | Patskovsky, Y, Madegowda, M, Gilmore, M, Chang, S, Maletic, M, Smith, D, Sauder, J.M, Burley, S.K, Swaminathan, S, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-29 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of a putative lactoylglutathione lyase from Bdellovibrio bacteriovorus
To be Published
|
|
3FYF
 
 | | Crystal structure of uncharacterized protein bvu_3222 from bacteroides vulgatus | | Descriptor: | PROTEIN BVU-3222 | | Authors: | Patskovsky, Y, Bonanno, J.B, Ozyurt, S, Rutter, M, Chang, S, Groshong, C, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-22 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Protein Bvu-3222 from Bacteroides Vulgatus
To be Published
|
|
6LK0
 
 | | Crystal structure of human wild type TRIP13 | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Wang, Y, Huang, J, Li, B, Xue, H, Tricot, G, Hu, L, Xu, Z, Sun, X, Chang, S, Gao, L, Tao, Y, Xu, H, Xie, Y, Xiao, W, Yu, D, Kong, Y, Chen, G, Sun, X, Lian, F, Zhang, N, Wu, X, Mao, Z, Zhan, F, Zhu, W, Shi, J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Small-Molecule Inhibitor Targeting TRIP13 Suppresses Multiple Myeloma Progression.
Cancer Res., 80, 2020
|
|
6VGI
 
 | | Crystal Structures of FLAP bound to MK-866 | | Descriptor: | 3-[3-(tert-butylsulfanyl)-1-[(4-chlorophenyl)methyl]-5-(propan-2-yl)-1H-indol-2-yl]-2,2-dimethylpropanoic acid, 5-lipoxygenase-activating protein, SULFATE ION | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6VGC
 
 | | Crystal Structures of FLAP bound to DG-031 | | Descriptor: | (2R)-cyclopentyl{4-[(quinolin-2-yl)methoxy]phenyl}acetic acid, 5-lipoxygenase-activating protein, CALCIUM ION, ... | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
2B1M
 
 | | Crystal structure of a papain-fold protein without the catalytic cysteine from seeds of Pachyrhizus erosus | | Descriptor: | DI(HYDROXYETHYL)ETHER, SPE31, TETRAETHYLENE GLYCOL, ... | | Authors: | Zhang, M, Wei, Z, Chang, S. | | Deposit date: | 2005-09-16 | | Release date: | 2006-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a papain-fold protein without the catalytic residue: a novel member in the cysteine proteinase family
J.Mol.Biol., 358, 2006
|
|
3C19
 
 | | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein MK0293 | | Authors: | Patskovsky, Y, Romero, R, Bonanno, J.B, Malashkevich, V, Dickey, M, Chang, S, Koss, J, Bain, K, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-22 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19.
To be Published
|
|
3C3M
 
 | | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1 | | Descriptor: | GLYCEROL, Response regulator receiver protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Dickey, M, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1.
To be Published
|
|
3CG0
 
 | | Crystal structure of signal receiver domain of modulated diguanylate cyclase from Desulfovibrio desulfuricans G20, an example of alternate folding | | Descriptor: | Response regulator receiver modulated diguanylate cyclase with PAS/PAC sensor | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Gilmore, M, Chang, S, Groshong, C, Koss, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Modulated Diguanylate Cyclase from Desulfovibrio desulfuricans.
To be Published
|
|
3EZY
 
 | | Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Dehydrogenase | | Authors: | Ramagopal, U.A, Toro, R, Freeman, J, Chang, S, Maletic, M, Gheyi, T, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-24 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima
To be published
|
|
3CZ8
 
 | | Crystal structure of putative sporulation-specific glycosylase ydhD from Bacillus subtilis | | Descriptor: | GLYCEROL, Putative sporulation-specific glycosylase ydhD | | Authors: | Patskovsky, Y, Romero, R, Rutter, M, Chang, S, Maletic, M, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-28 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative glycosylase ydhD from Bacillus subtilis.
To be Published
|
|
3CRN
 
 | | Crystal structure of response regulator receiver domain protein (CheY-like) from Methanospirillum hungatei JF-1 | | Descriptor: | GLYCEROL, Response regulator receiver domain protein, CheY-like, ... | | Authors: | Hiner, R.L, Toro, R, Patskovsky, Y, Freeman, J, Chang, S, Smith, D, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-07 | | Release date: | 2008-04-22 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of response regulator receiver domain protein (CheY-like) from Methanospirillum hungatei JF-1.
To be Published
|
|
7XUO
 
 | | Structure of ATP7B C983S/C985S/D1027A mutant with cisplatin in presence of ATOX1 | | Descriptor: | Copper-transporting ATPase 2, PLATINUM (II) ION | | Authors: | Yang, G, Xu, L, Chang, S, Guo, J, Wu, Z. | | Deposit date: | 2022-05-19 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
