2M2Q
 
 | | Solution structure of MCh-1: A novel inhibitor cystine knot peptide from Momordica charantia | | Descriptor: | Inhibitor cystine knot peptide MCh-1 | | Authors: | He, W, Chan, L, Clark, R.J, Tang, J, Zeng, G, Franco, O.L, Cantacessi, C, Craik, D.J, Daly, N.L, Tan, N. | | Deposit date: | 2013-01-01 | | Release date: | 2013-11-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Novel Inhibitor Cystine Knot Peptides from Momordica charantia.
Plos One, 8, 2013
|
|
2ZKU
 
 | | Structure of hepatitis C virus NS5B polymerase in a new crystal form | | Descriptor: | ACETIC ACID, GLYCEROL, Genome polyprotein | | Authors: | Biswal, B.K. | | Deposit date: | 2008-03-31 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Hepatitis C Virus Ns5B Polymerase in a New Crystal Form: Insights Into Oligomerisation and Allosteric Nucleotide Binding Site
To be Published
|
|
2HXA
 
 | |
2HX9
 
 | |
2HX7
 
 | |
2HX8
 
 | |
4PZX
 
 | | Synthesis, Characterization and PK/PD Studies of a Series of Spirocyclic Pyranochromene BACE1 Inhibitors | | Descriptor: | (4R,4a'R,10a'R)-7'-(5-chloropyridin-3-yl)-3',4',4a',10a'-tetrahydro-1'H-spiro[1,3-oxazole-4,5'-pyrano[3,4-b]chromen]-2-amine, Beta-secretase 1, NICKEL (II) ION | | Authors: | Vigers, G.P.A. | | Deposit date: | 2014-03-31 | | Release date: | 2014-05-14 | | Last modified: | 2014-10-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, characterization, and PK/PD studies of a series of spirocyclic pyranochromene BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4PZW
 
 | | Synthesis, Characterization and PK/PD Studies of a Series of Spirocyclic Pyranochromene BACE1 Inhibitors | | Descriptor: | (4R,4a'S,10a'S)-7'-(5-chloropyridin-3-yl)-3',4',4a',10a'-tetrahydro-1'H-spiro[1,3-oxazole-4,5'-pyrano[3,4-b]chromen]-2-amine, Beta-secretase 1, NICKEL (II) ION | | Authors: | Vigers, G.P.A. | | Deposit date: | 2014-03-31 | | Release date: | 2014-05-14 | | Last modified: | 2014-10-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, characterization, and PK/PD studies of a series of spirocyclic pyranochromene BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6XF5
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (RBDs down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
6XF6
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
1M6O
 
 | | Crystal Structure of HLA B*4402 in complex with HLA DPA*0201 peptide | | Descriptor: | Beta-2-microglobulin, HLA DPA*0201 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Macdonald, W.A, Purcell, A.W, Williams, D.S, Mifsud, N.A, Ely, L.K, Gorman, J.J, Clements, C.S, Kjer-Nielsen, L, Koelle, D.M, Brooks, A.G, Lovrecz, G.O, Lu, L, Rossjohn, J, McCluskey, J. | | Deposit date: | 2002-07-17 | | Release date: | 2003-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition.
J.Exp.Med., 198, 2003
|
|
1N2R
 
 | | A natural selected dimorphism in HLA B*44 alters self, peptide reportoire and T cell recognition. | | Descriptor: | ACETIC ACID, Beta-2-microglobulin, HLA DPA*0201 PEPTIDE, ... | | Authors: | Macdonald, W.A, Purcell, A.W, Williams, D.S, Mifsud, N, Ely, L.K, Gorman, J.J, Clements, C.S, Kjer-Nielsen, L, Koelle, D.M, Brooks, A.G, Lovrecz, G.O, Lu, L, Rossjohn, J, McCluskey, J. | | Deposit date: | 2002-10-24 | | Release date: | 2004-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition.
J.Exp.Med., 198, 2003
|
|
6X58
 
 | | MPER-Fluc-Ec2 bound to 10E8v4 antibody | | Descriptor: | 10E8v4 Fab Heavy Chain, 10E8v4 Fab Light Chain, gp41 MPER peptide,Putative fluoride ion transporter CrcB | | Authors: | McIlwain, B.C, Stockbridge, R.B. | | Deposit date: | 2020-05-25 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | N-terminal Transmembrane-Helix Epitope Tag for X-ray Crystallography and Electron Microscopy of Small Membrane Proteins.
J.Mol.Biol., 433, 2021
|
|
2GJJ
 
 | | Crystal structure of a single chain antibody scA21 against Her2/ErbB2 | | Descriptor: | A21 single-chain antibody fragment against erbB2, GLYCEROL | | Authors: | Zhu, Z. | | Deposit date: | 2006-03-31 | | Release date: | 2006-10-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Epitope mapping and structural analysis of an anti-ErbB2 antibody A21: Molecular basis for tumor inhibitory mechanism
Proteins, 70, 2007
|
|
4Y8D
 
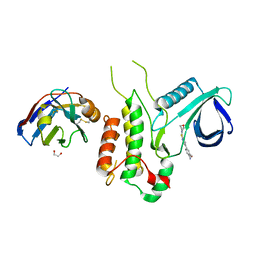 | | Crystal structure of Cyclin-G associated kinase (GAK) complexed with selective 12i inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-[3-(morpholin-4-yl)[1,2]thiazolo[4,3-b]pyridin-6-yl]aniline, Cyclin-G-associated kinase, ... | | Authors: | Chaikuad, A, Heroven, C, Nowak, R, De Jonghe, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-16 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective Inhibitors of Cyclin G Associated Kinase (GAK) as Anti-Hepatitis C Agents.
J.Med.Chem., 58, 2015
|
|
7LUC
 
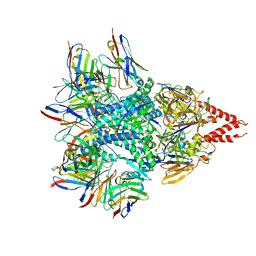 | | Cryo-EM structure of RSV preF bound by Fabs 32.4K and 01.4B | | Descriptor: | 01.4B Fab Heavy chain, 01.4B Fab Light chain, 32.4K Fab Heavy chain, ... | | Authors: | Wrapp, D, McLellan, J.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-21 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Vaccination with prefusion-stabilized respiratory syncytial virus fusion protein induces genetically and antigenically diverse antibody responses.
Immunity, 54, 2021
|
|
7LUE
 
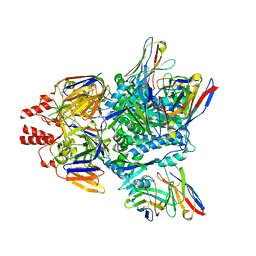 | |
7LUD
 
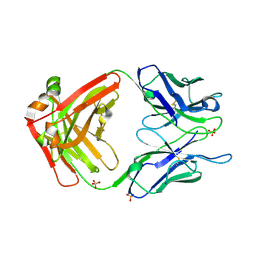 | | Crystal structure of Fab ADI-14442 | | Descriptor: | ADI-14442 Fab Heavy chain, ADI-14442 Fab Light chain, SULFATE ION | | Authors: | Rush, S.A, McLellan, J.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Vaccination with prefusion-stabilized respiratory syncytial virus fusion protein induces genetically and antigenically diverse antibody responses.
Immunity, 54, 2021
|
|
5LGG
 
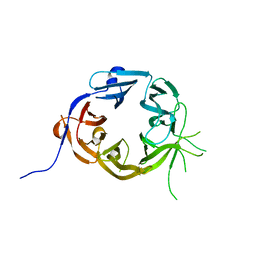 | | The N-terminal WD40 domain of Apc1 (Anaphase promoting complex subunit 1) | | Descriptor: | Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1 | | Authors: | Li, Q, Aibara, S, Barford, D. | | Deposit date: | 2016-07-07 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | WD40 domain of Apc1 is critical for the coactivator-induced allosteric transition that stimulates APC/C catalytic activity.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5W5V
 
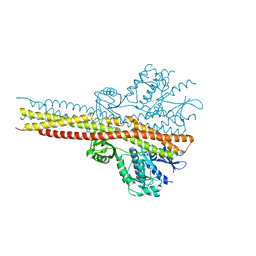 | | TBK1 co-crystal structure with amlexanox | | Descriptor: | 2-amino-7-(1-methylethyl)-5-oxo-5H-chromeno[2,3-b]pyridine-3-carboxylic acid, Serine/threonine-protein kinase TBK1 | | Authors: | Beyett, T.S, Tesmer, J.J.G. | | Deposit date: | 2017-06-15 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.645 Å) | | Cite: | Carboxylic Acid Derivatives of Amlexanox Display Enhanced Potency toward TBK1 and IKKepsilonand Reveal Mechanisms for Selective Inhibition.
Mol. Pharmacol., 94, 2018
|
|
5TXF
 
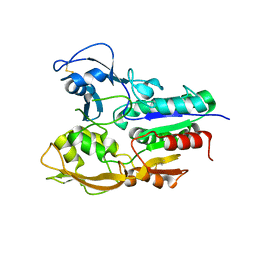 | | Crystal structure of Lecithin:cholesterol acyltransferase (LCAT) in a closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Phosphatidylcholine-sterol acyltransferase, ... | | Authors: | Manthei, K.A, Glukhova, A, Tesmer, J.J.G. | | Deposit date: | 2016-11-16 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A retractable lid in lecithin:cholesterol acyltransferase provides a structural mechanism for activation by apolipoprotein A-I.
J. Biol. Chem., 292, 2017
|
|
5DJ4
 
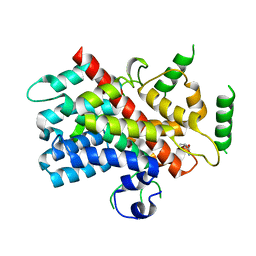 | |
6CQ4
 
 | | TBK1 in Complex with Cyclohexyl Analog of Amlexanox | | Descriptor: | 2-amino-7-(4,4-difluorocyclohexyl)-5-oxo-5H-[1]benzopyrano[2,3-b]pyridine-3-carboxylic acid, Serine/threonine-protein kinase TBK1 | | Authors: | Beyett, T.S, Tesmer, J.J.G. | | Deposit date: | 2018-03-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design, synthesis, and biological activity of substituted 2-amino-5-oxo-5H-chromeno[2,3-b]pyridine-3-carboxylic acid derivatives as inhibitors of the inflammatory kinases TBK1 and IKK epsilon for the treatment of obesity.
Bioorg. Med. Chem., 26, 2018
|
|
6CES
 
 | | Cryo-EM structure of GATOR1-RAG | | Descriptor: | GATOR complex protein DEPDC5, GATOR complex protein NPRL2, GATOR complex protein NPRL3, ... | | Authors: | Shen, K, Huang, R.K, Brignole, E.J, Yu, Z, Sabatini, D.M. | | Deposit date: | 2018-02-12 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes.
Nature, 556, 2018
|
|
6CQ0
 
 | | TBK1 in Complex with Dimethyl Amino Analog of Amlexanox | | Descriptor: | 2-amino-7-[3-(dimethylamino)propyl]-5-oxo-5H-[1]benzopyrano[2,3-b]pyridine-3-carboxylic acid, Serine/threonine-protein kinase TBK1 | | Authors: | Beyett, T.S, Tesmer, J.J.G. | | Deposit date: | 2018-03-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Design, synthesis, and biological activity of substituted 2-amino-5-oxo-5H-chromeno[2,3-b]pyridine-3-carboxylic acid derivatives as inhibitors of the inflammatory kinases TBK1 and IKK epsilon for the treatment of obesity.
Bioorg. Med. Chem., 26, 2018
|
|
