3C2X
 
 | | Crystal structure of peptidoglycan recognition protein at 1.8A resolution | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Peptidoglycan recognition protein, ... | | Authors: | Sharma, P, Singh, N, Sinha, M, Sharma, S, Perbandt, M, Betzel, C, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2008-01-26 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the peptidoglycan recognition protein at 1.8 A resolution reveals dual strategy to combat infection through two independent functional homodimers
J.Mol.Biol., 378, 2008
|
|
7PXZ
 
 | | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
7PZQ
 
 | | Oxidized form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
1FV0
 
 | | FIRST STRUCTURAL EVIDENCE OF THE INHIBITION OF PHOSPHOLIPASE A2 BY ARISTOLOCHIC ACID: CRYSTAL STRUCTURE OF A COMPLEX FORMED BETWEEN PHOSPHOLIPASE A2 AND ARISTOLOCHIC ACID | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 9-HYDROXY ARISTOLOCHIC ACID, ACETATE ION, ... | | Authors: | Chandra, V, Jasti, J, Kaur, P, Srinivasan, A, Betzel, C, Singh, T.P. | | Deposit date: | 2000-09-18 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Phospholipase A2 Inhibition for the Synthesis of Prostaglandins by the Plant Alkaloid Aristolochic Acid from a 1.7 A Crystal Structure
Biochemistry, 41, 2002
|
|
7Q0Y
 
 | | Crystal structure of CTX-M-14 in complex with Bortezomib | | Descriptor: | ACETATE ION, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Werner, N, Perbandt, M, Hinrichs, W, Prester, A, Rohde, H, Aepfelbacher, M, Betzel, C. | | Deposit date: | 2021-10-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as beta-lactamase inhibitors.
Sci Rep, 12, 2022
|
|
7Q11
 
 | | Crystal structure of CTX-M-14 in complex with Ixazomib | | Descriptor: | Beta-lactamase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Werner, N, Perbandt, M, Hinrichs, W, Prester, A, Rohde, H, Aepfelbacher, M, Betzel, C. | | Deposit date: | 2021-10-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as beta-lactamase inhibitors.
Sci Rep, 12, 2022
|
|
7Q0Z
 
 | | Crystal structure of CTX-M-14 | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Werner, N, Perbandt, M, Hinrichs, W, Prester, A, Rohde, H, Aepfelbacher, M, Betzel, C. | | Deposit date: | 2021-10-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as beta-lactamase inhibitors.
Sci Rep, 12, 2022
|
|
1EMR
 
 | | CRYSTAL STRUCTURE OF HUMAN LEUKEMIA INHIBITORY FACTOR (LIF) | | Descriptor: | LEUKEMIA INHIBITORY FACTOR | | Authors: | Robinson, R.C, Heath, J.K, Hawkins, N, Samal, B, Jones, E.Y, Betzel, C. | | Deposit date: | 2000-03-17 | | Release date: | 2001-03-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Species Variation in Receptor Binding Site Revealed by the Medium Resolution X-ray Structure of Human Leukemia Inhibitory Factor
to be published
|
|
7QCG
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2-pyrrolidyl)-3,4,5-trihydroxybenzoylhydrazone | | Descriptor: | 3,4,5-tris(oxidanyl)-N-[(E)-1H-pyrrol-2-ylmethylideneamino]benzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-23 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCM
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3-methoxy-4-hydroxy-acetophenone)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3-metoxy-4-hydroxy-acetophenone)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCJ
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCK
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,5-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,5-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCI
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCH
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,5-dimethoxy-4-hydroxybenzyliden)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,5-dimetoxy-4-hydroxybenzyliden)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
2TEC
 
 | | MOLECULAR DYNAMICS REFINEMENT OF A THERMITASE-EGLIN-C COMPLEX AT 1.98 ANGSTROMS RESOLUTION AND COMPARISON OF TWO CRYSTAL FORMS THAT DIFFER IN CALCIUM CONTENT | | Descriptor: | CALCIUM ION, EGLIN C, THERMITASE | | Authors: | Gros, P, Betzel, C, Dauter, Z, Wilson, K.S, Hol, W.G.J. | | Deposit date: | 1990-10-26 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular dynamics refinement of a thermitase-eglin-c complex at 1.98 A resolution and comparison of two crystal forms that differ in calcium content.
J.Mol.Biol., 210, 1989
|
|
1NTN
 
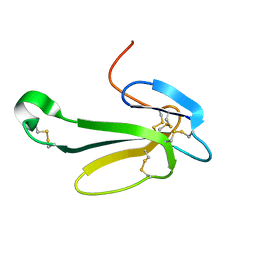 | | THE CRYSTAL STRUCTURE OF NEUROTOXIN-I FROM NAJA NAJA OXIANA AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | NEUROTOXIN I | | Authors: | Mikhailov, A.M, Nickitenko, A.V, Vainshtein, B.K, Betzel, C, Wilson, K. | | Deposit date: | 1994-09-26 | | Release date: | 1995-05-08 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-dimensional structure of neurotoxin-1 from Naja naja oxiana venom at 1.9 A resolution.
Febs Lett., 320, 1993
|
|
1LNL
 
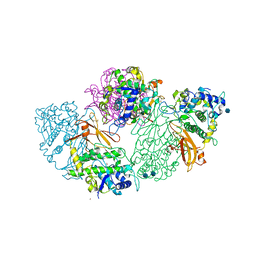 | | Structure of deoxygenated hemocyanin from Rapana thomasiana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, hemocyanin | | Authors: | Perbandt, M, Guthoehrlein, E.W, Rypniewski, W, Idakieva, K, Stoeva, S, Voelter, W, Genov, N, Betzel, C. | | Deposit date: | 2002-05-03 | | Release date: | 2003-06-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of a functional unit from the wall of a gastropod hemocyanin offers a possible mechanism for cooperativity
Biochemistry, 42, 2003
|
|
4FYU
 
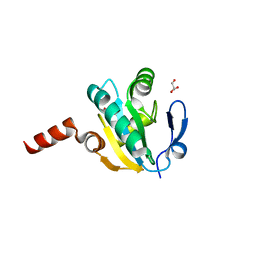 | | Crystal structure of Thioredoxin from Wuchereria bancrofti at 2.0 Angstrom | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Thioredoxin | | Authors: | Yousef, N, Prabhu, P, Ahmed, A, Iqbal, S, Betzel, C. | | Deposit date: | 2012-07-05 | | Release date: | 2013-08-07 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Thioredoxin from Wuchereria bancrofti at 2.0 Angstrom
TO BE PUBLISHED
|
|
4G8J
 
 | | X-ray Structure of Uridine Phosphorylease from Vibrio cholerae Complexed with Thymidine at 2.12 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Lashkov, A.A, Gabdoulkhakov, A.G, Prokofev, I.I, Sotnichenko, S.E, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2012-07-23 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | X-ray structure of uridine phosphorylease from Vibrio cholerae complexed with thymidine
To be Published
|
|
4IP0
 
 | | X-Ray Structure of the Complex Uridine Phosphorylase from Vibrio cholerae with Phosphate Ion at 1.29 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ETHANOL, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2013-01-09 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.294 Å) | | Cite: | X-ray structures of uridine phosphorylase from Vibrio cholerae in complexes with uridine, thymidine, uracil, thymine, and phosphate anion: Substrate specificity of bacterial uridine phosphorylases
Crystallography Reports, 61, 2016
|
|
4GY7
 
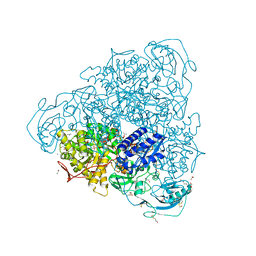 | | Crystallographic structure analysis of urease from Jack bean (Canavalia ensiformis) at 1.49 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETONE, BETA-MERCAPTOETHANOL, ... | | Authors: | Begum, A, Banumathi, S, Choudhary, M.I, Betzel, C. | | Deposit date: | 2012-09-05 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | Crystallographic structure analysis of urease from Jack bean (Canavalia ensiformis) at 1.49 A Resolution
To be Published
|
|
4H1T
 
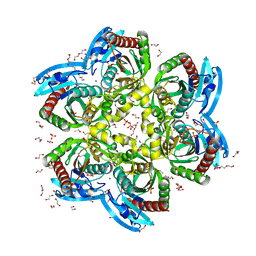 | | X-RAY Structure of the Complex VchUPh with Phosphate ion at 1.92A Resolution. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Sotnichenko, S.E, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2012-09-11 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.924 Å) | | Cite: | X-RAY Structure of the Complex VchUPh with Phosphate ion at 1.92A Resolution.
TO BE PUBLISHED
|
|
4I2V
 
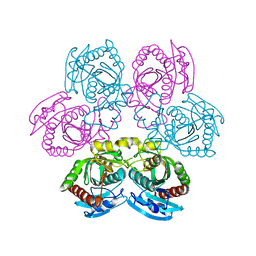 | | X-ray structure of the unliganded uridine phosphorylase from Yersinia pseudotuberculosis at 2.12A resolution | | Descriptor: | Uridine phosphorylase | | Authors: | Lashkov, A.A, Balaev, V.V, Prokofev, I.I, Betzel, C, Gabdoulkhakov, A.G, Mikhailov, A.M. | | Deposit date: | 2012-11-23 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | X-ray structure of the unliganded uridine phosphorylase from Yersinia pseudotuberculosis at 2.12A resolution
To be Published
|
|
4GV5
 
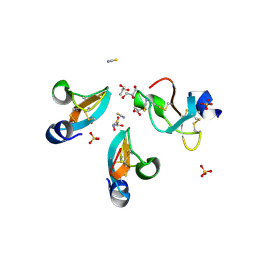 | | X-ray structure of crotamine, a cell-penetrating peptide from the Brazilian snake Crotalus durissus terrificus | | Descriptor: | Crotamine Ile-19, GLYCEROL, SULFATE ION, ... | | Authors: | Coronado, M.A, Gabdulkhakov, A, Georgieva, D, Sankaran, B, Murakami, M.T, Arni, R.K, Betzel, C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the polypeptide crotamine from the Brazilian rattlesnake Crotalus durissus terrificus.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4JP5
 
 | | X-ray structure of uridine phosphorylase from Yersinia pseudotuberculosis in unliganded state at 2.27 A resolution | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Prokofev, I.I, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2013-03-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | X-ray structure of uridine phosphorylase from Yersinia pseudotuberculosis in unliganded state at 2.27 A resolution
To be Published
|
|
