7FAU
 
 | | Structure Determination of the NB1B11-RBD Complex | | Descriptor: | NB_1B11, Spike protein S1, ZINC ION | | Authors: | Shi, Z.Z, Li, X.X, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C, Cui, Q.Q, Qiao, H.R, Lan, Z.Y, Zhang, X. | | Deposit date: | 2021-07-07 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of nanobodies neutralizing SARS-CoV-2 variants.
Structure, 30, 2022
|
|
3CIB
 
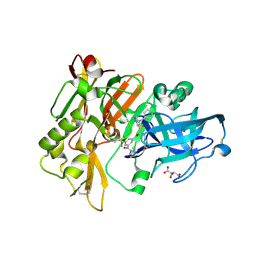 | | Structure of BACE Bound to SCH727596 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2R)-2-[(2R,4S)-4-benzylpiperidin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4FA8
 
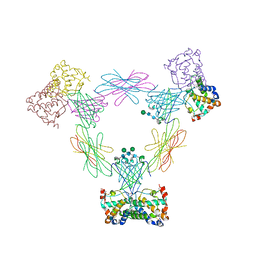 | | Multi-pronged modulation of cytokine signaling | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage colony-stimulating factor 1, Secreted protein BARF1, ... | | Authors: | He, X, Shim, A.H. | | Deposit date: | 2012-05-21 | | Release date: | 2012-08-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multipronged attenuation of macrophage-colony stimulating factor signaling by Epstein-Barr virus BARF1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8JOT
 
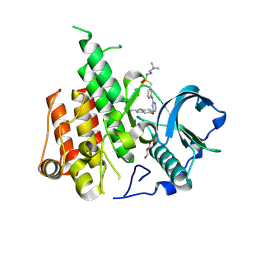 | | Crystal structure of CSF-1R kinase domain with sulfatinib | | Descriptor: | GLYCEROL, Macrophage colony-stimulating factor 1 receptor, N-(2-(dimethylamino)ethyl)-1-(3-((4-((2-methyl-1H-indol-5-yl)oxy)pyrimidin-2-yl)amino)phenyl)methanesulfonamide | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis and selectivity of sulfatinib binding to FGFR and CSF-1R.
Commun Chem, 7, 2024
|
|
6JFK
 
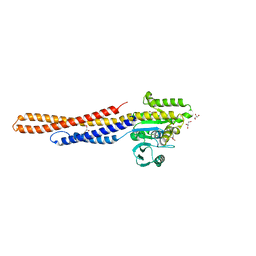 | | GDP bound Mitofusin2 (MFN2) | | Descriptor: | CITRIC ACID, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Li, Y.J, Cao, Y.L, Feng, J.X, Qi, Y.B, Meng, S.X, Yang, J.F, Zhong, Y.T, Kang, S.S, Chen, X.X, Lan, L, Luo, L, Yu, B, Chen, S.D, Chan, D.C, Hu, J.J, Gao, S. | | Deposit date: | 2019-02-10 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural insights of human mitofusin-2 into mitochondrial fusion and CMT2A onset.
Nat Commun, 10, 2019
|
|
7F3M
 
 | | Crystal structure of FGFR4 kinase domain with PRN1371 | | Descriptor: | 6-[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]-2-(methylamino)-8-[3-(4-prop-2-enoylpiperazin-1-yl)propyl]pyrido[2,3-d]pyrimidin-7-one, Fibroblast growth factor receptor 4, SULFATE ION | | Authors: | Chen, X.J, Qu, L.Z, Dai, S.Y, Wei, H.D, Chen, Y.H. | | Deposit date: | 2021-06-16 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
6UG1
 
 | | Sequence impact in DNA duplex opening by the Rad4/XPC nucleotide excision repair complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*GP*GP*AP*TP*GP*TP*CP*GP*AP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*(G47)P*AP*CP*AP*TP*CP*CP*CP*CP*TP*AP*CP*AP*A)-3'), DNA repair protein RAD4, ... | | Authors: | Paul, D, Min, J.-H. | | Deposit date: | 2019-09-25 | | Release date: | 2021-03-31 | | Last modified: | 2021-09-08 | | Method: | X-RAY DIFFRACTION (2.833 Å) | | Cite: | Impact of DNA sequences on DNA 'opening' by the Rad4/XPC nucleotide excision repair complex.
DNA Repair (Amst), 107, 2021
|
|
8W9A
 
 | | CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-7909 binding at an allosteric site | | Descriptor: | 6-chloranyl-3-[[(1R)-1-[2-(1,3-dihydropyrrolo[3,4-c]pyridin-2-yl)-3,6-dimethyl-4-oxidanylidene-quinazolin-8-yl]ethyl]amino]pyridine-2-carboxylic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Huang, X, Ren, X, Zhong, W. | | Deposit date: | 2023-09-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures reveal two allosteric inhibition modes of PI3K alpha H1047R involving a re-shaping of the activation loop.
Structure, 32, 2024
|
|
8W9B
 
 | | CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-8557 binding at an allosteric site | | Descriptor: | 1-[(1S)-1-(5-fluoranyl-3-methyl-1-benzofuran-2-yl)-2-methyl-propyl]-3-(1-oxidanylidene-2,3-dihydroisoindol-5-yl)urea, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Huang, X, Ren, X, Zhong, W. | | Deposit date: | 2023-09-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures reveal two allosteric inhibition modes of PI3K alpha H1047R involving a re-shaping of the activation loop.
Structure, 32, 2024
|
|
7FAT
 
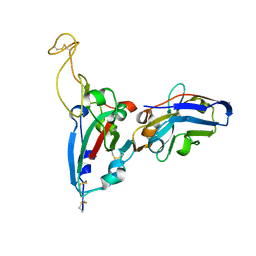 | | Structure Determination of the RBD-NB1A7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nb_1A7 | | Authors: | Geng, Y, Shi, Z.Z, Li, X.Y, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of nanobodies neutralizing SARS-CoV-2 variants
Structure, 30, 2022
|
|
5ZQZ
 
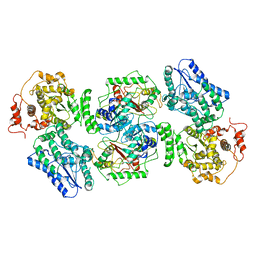 | | Structure of human mitochondrial trifunctional protein, tetramer | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | Deposit date: | 2018-04-20 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZRV
 
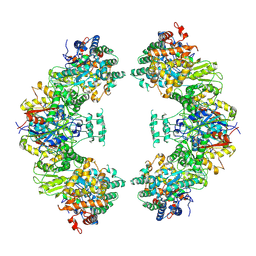 | | Structure of human mitochondrial trifunctional protein, octamer | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | Deposit date: | 2018-04-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7D5O
 
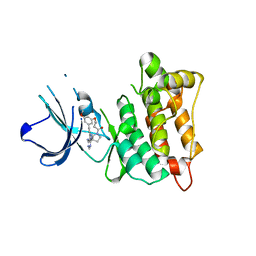 | | C-Src in complex with TAS-120 | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]prop-2-en-1-one, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2020-09-27 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
7DOQ
 
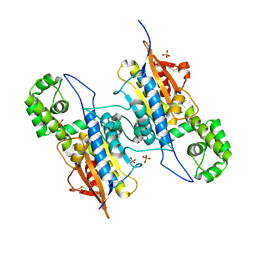 | | Lp major histidine acid phosphatase mutant D281A/5'-AMP | | Descriptor: | Acid phosphatase, PHOSPHATE ION | | Authors: | Guo, Y, Teng, Y. | | Deposit date: | 2020-12-16 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into a new substrate binding mode of a histidine acid phosphatase from Legionella pneumophila.
Biochem.Biophys.Res.Commun., 540, 2021
|
|
3G0Y
 
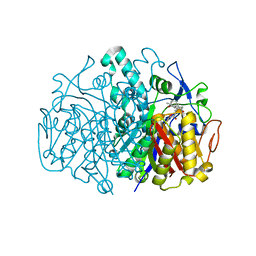 | | Structure of E. coli FabF(C163Q) in complex with dihydroplatensimycin | | Descriptor: | 3-({3-[(1S,4aS,6S,7S,9S,9aR)-1,6-dimethyl-2-oxodecahydro-6,9-epoxy-4a,7-methanobenzo[7]annulen-1-yl]propanoyl}amino)-2,4-dihydroxybenzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | Authors: | Soisson, S.M, Parthasarathy, G. | | Deposit date: | 2009-01-29 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis and biological evaluation of platensimycin analogs.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3G11
 
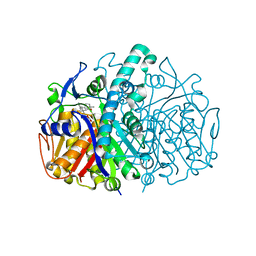 | | Structure of E. coli FabF(C163Q) in complex with dihydrophenyl platensimycin | | Descriptor: | 3-({3-[(1S,4S,4aS,6S,7S,9S,9aR)-1,6-dimethyl-2-oxo-4-phenyldecahydro-6,9-epoxy-4a,7-methanobenzo[7]annulen-1-yl]propanoyl}amino)-2,4-dihydroxybenzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | Authors: | Soisson, S.M, Parthasarathy, G. | | Deposit date: | 2009-01-29 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and biological evaluation of platensimycin analogs.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2F4B
 
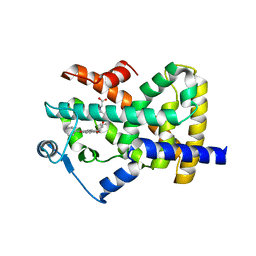 | | Crystal structure of the ligand binding domain of human PPAR-gamma in complex with an agonist | | Descriptor: | (5-{3-[(6-BENZOYL-1-PROPYL-2-NAPHTHYL)OXY]PROPOXY}-1H-INDOL-1-YL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Lu, I.L, Peng, Y.H, Mahindroo, N, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2005-11-23 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Indol-1-yl Acetic Acids as Peroxisome Proliferator-Activated Receptor Agonists: Design, Synthesis, Structural Biology, and Molecular Docking Studies
J.Med.Chem., 49, 2006
|
|
4H3F
 
 | | Structure of BACE Bound to 3-(5-((7aR)-2-imino-6-(6-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-1H-pyrrolo[3,4-d]pyrimidin-7a-yl)thiophen-3-yl)benzonitrile | | Descriptor: | 3-{5-[(2E,4aR,7aR)-2-imino-6-(6-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-7aH-pyrrolo[3,4-d]pyrimidin-7a-yl]thiophen-3-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4H3G
 
 | | Structure of BACE Bound to 2-((7aR)-7a-(4-(3-cyanophenyl)thiophen-2-yl)-2-imino-3-methyl-4-oxohexahydro-1H-pyrrolo[3,4-d]pyrimidin-6(2H)-yl)nicotinonitrile | | Descriptor: | 2-{(2E,4aR,7aR)-7a-[4-(3-cyanophenyl)thiophen-2-yl]-2-imino-3-methyl-4-oxooctahydro-6H-pyrrolo[3,4-d]pyrimidin-6-yl}pyridine-3-carbonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4HA5
 
 | | Structure of BACE Bound to (S)-3-(5-(2-imino-1,4-dimethyl-6-oxohexahydropyrimidin-4-yl)thiophen-3-yl)benzonitrile | | Descriptor: | 3-{5-[(2E,4S)-2-imino-1,4-dimethyl-6-oxohexahydropyrimidin-4-yl]thiophen-3-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-25 | | Release date: | 2012-10-17 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4H3J
 
 | | Structure of BACE Bound to 2-fluoro-5-(5-(2-imino-3-methyl-4-oxo-6-phenyloctahydro-1H-pyrrolo[3,4-d]pyrimidin-7a-yl)thiophen-2-yl)benzonitrile | | Descriptor: | 2-fluoro-5-{5-[(2E,4aR,7aR)-2-imino-3-methyl-4-oxo-6-phenyloctahydro-7aH-pyrrolo[3,4-d]pyrimidin-7a-yl]thiophen-2-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-10-17 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
8HPB
 
 | |
8HQB
 
 | | NMR Structure of OsCIE1-Ubox | | Descriptor: | U-box domain-containing protein 12 | | Authors: | Zhang, Y, Yu, C.Z, Lan, W.X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Release of a ubiquitin brake activates OsCERK1-triggered immunity in rice.
Nature, 629, 2024
|
|
4H1E
 
 | |
4H3I
 
 | | Structure of BACE Bound to 3-(5-((7aR)-2-imino-6-(3-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-1H-pyrrolo[3,4-d]pyrimidin-7a-yl)thiophen-3-yl)benzonitrile | | Descriptor: | 3-{5-[(2E,4aR,7aR)-2-imino-6-(3-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-7aH-pyrrolo[3,4-d]pyrimidin-7a-yl]thiophen-3-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
