1VM2
 
 | |
1VM4
 
 | |
8X43
 
 | | human KCNQ2-CaM-Ebio1-S1 complex in the presence of PIP2 | | Descriptor: | Calmodulin-1, N-(4-azanyl-1,2-dihydroacenaphthylen-5-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2023-11-15 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A small-molecule activation mechanism that directly opens the KCNQ2 channel.
Nat.Chem.Biol., 20, 2024
|
|
7JM5
 
 | | Crystal structure of KDM4B in complex with QC6352 | | Descriptor: | 3-[({(1R)-6-[methyl(phenyl)amino]-1,2,3,4-tetrahydronaphthalen-1-yl}methyl)amino]pyridine-4-carboxylic acid, Lysine-specific demethylase 4B, NICKEL (II) ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-07-31 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting KDM4 for treating PAX3-FOXO1-driven alveolar rhabdomyosarcoma.
Sci Transl Med, 14, 2022
|
|
6AN6
 
 | | Crystal structure of Escherichia coli HPPK in complex with bisubstrate analogue inhibitor HP-72 | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-{2-[{2-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carbonyl)amino]ethyl}(phosphonomethyl)amino]ethyl}-5'-thioadenosine | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2017-08-12 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bisubstrate analogue inhibitors of HPPK: Transition state mimetics
to be published
|
|
1IHT
 
 | |
1IHS
 
 | |
4QR5
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | Bromodomain-containing protein 4, N-[3-(cyclopentylsulfamoyl)-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]cyclopropanecarboxamide | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
2MVT
 
 | | Solution structure of scoloptoxin SSD609 from Scolopendra mutilans | | Descriptor: | Scoloptoxin SSD609 | | Authors: | Wu, F, Sun, P, Wang, C, He, Y, Zhang, L, Tian, C. | | Deposit date: | 2014-10-14 | | Release date: | 2015-09-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A distinct three-helix centipede toxin SSD609 inhibits Iks channels by interacting with the KCNE1 auxiliary subunit.
Sci Rep, 5, 2015
|
|
4QR3
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | Bromodomain-containing protein 4, N-cyclopentyl-3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
7F5G
 
 | | The crystal structure of RBD-Nanobody complex, DL4 (SA4) | | Descriptor: | ACETATE ION, GLYCEROL, Nanobody DL4, ... | | Authors: | Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isolation, characterization, and structure-based engineering of a neutralizing nanobody against SARS-CoV-2.
Int.J.Biol.Macromol., 209, 2022
|
|
8H1T
 
 | | Cryo-EM structure of BAP1-ASXL1 bound to chromatosome | | Descriptor: | DNA (187-MER), Histone H1.4, Histone H2A type 1-D, ... | | Authors: | Ge, W, Yu, C, Xu, R.M. | | Deposit date: | 2022-10-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Basis of the H2AK119 specificity of the Polycomb repressive deubiquitinase.
Nature, 616, 2023
|
|
4QR4
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | 2-chloro-N-cyclopentyl-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
7F5H
 
 | | The crystal structure of RBD-Nanobody complex, DL28 (SC4) | | Descriptor: | GLYCEROL, Nanobody DL28, PHOSPHATE ION, ... | | Authors: | Luo, Z.P, Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Characterization of a Neutralizing Nanobody With Broad Activity Against SARS-CoV-2 Variants.
Front Microbiol, 13, 2022
|
|
8H7Z
 
 | |
8H7L
 
 | | Cryo-EM Structure of SARS-CoV-2 BA.2 Spike protein in complex with BA7535 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7535 fab heavt chain, ... | | Authors: | Liu, Z, Yan, A, Gao, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Identification of a highly conserved neutralizing epitope within the RBD region of diverse SARS-CoV-2 variants.
Nat Commun, 15, 2024
|
|
6BPH
 
 | | Crystal structure of the chromodomain of RBBP1 | | Descriptor: | AT-rich interactive domain-containing protein 4A, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-23 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of chromo barrel domain of RBBP1.
Biochem. Biophys. Res. Commun., 496, 2018
|
|
7JYA
 
 | | Crystal structure of E3 ligase in complex with peptide | | Descriptor: | ASN-ARG-ARG-ARG-ARG-TRP-ARG-GLU-ARG-GLN-ARG, Protein fem-1 homolog C, UNKNOWN ATOM OR ION | | Authors: | Yan, X, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Dong, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-30 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Molecular basis for ubiquitin ligase CRL2 FEM1C -mediated recognition of C-degron.
Nat.Chem.Biol., 17, 2021
|
|
7EGG
 
 | | TFIID lobe B subcomplex | | Descriptor: | Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 12, Transcription initiation factor TFIID subunit 4, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGF
 
 | | TFIID lobe A subcomplex | | Descriptor: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 11, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 2021
|
|
7EGH
 
 | | TFIID lobe C subcomplex | | Descriptor: | DNA (45-MER), Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 2, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGI
 
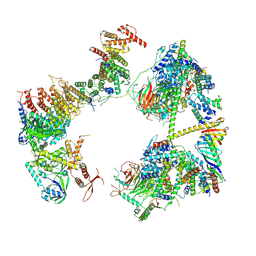 | | TFIID in rearranged conformation | | Descriptor: | TATA-box-binding protein, Transcription initiation factor IIA subunit 1, Transcription initiation factor IIA subunit 2, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (9.82 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGD
 
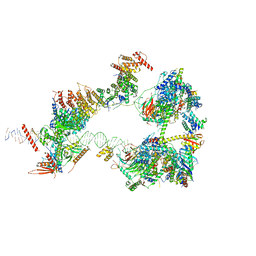 | | SCP promoter-bound TFIID-TFIIA in initial TBP-loading state | | Descriptor: | DNA (72-MER), TATA-box-binding protein, Transcription initiation factor IIA subunit 1, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (6.75 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGJ
 
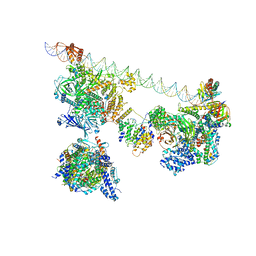 | | SCP promoter-bound TFIID-TFIIA in post TBP-loading state | | Descriptor: | DNA (74-MER), TATA-box-binding protein, Transcription initiation factor IIA subunit 1, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (8.64 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGE
 
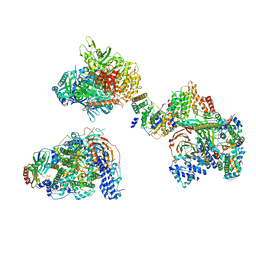 | | TFIID in canonical conformation | | Descriptor: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 10, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
