4HXL
 
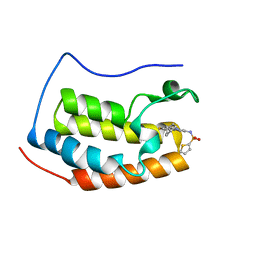 | | Brd4 Bromodomain 1 complex with 3-CYCLOHEXYL-N-{3-(2-OXO-2,3-DIHYDRO-1,3-THIAZOL-4-YL)-5-[(THIOPHEN-2-YLSULFONYL)AMINO]PHENYL}PROPANAMIDE inhibitor | | Descriptor: | 3-cyclohexyl-N-{3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)-5-[(thiophen-2-ylsulfonyl)amino]phenyl}propanamide, Bromodomain-containing protein 4 | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4ODE
 
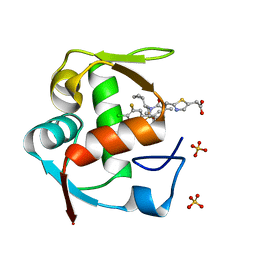 | | Co-Crystal Structure of MDM2 with Inhibitor Compound 4 | | Descriptor: | (2-{[(3R,5R,6S)-1-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(4-chloro-3-fluorophenyl)-5-(3-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]methyl}-1,3-thiazol-5-yl)acetic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
5HJS
 
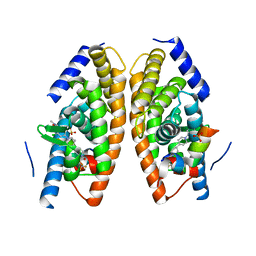 | | Identification of LXRbeta selective agonists for the treatment of Alzheimer's Disease | | Descriptor: | 2-chloro-4-{1'-[(2R)-2-hydroxy-3-methyl-2-(trifluoromethyl)butanoyl]-4,4'-bipiperidin-1-yl}-N,N-dimethylbenzamide, Nuclear receptor coactivator 1, Oxysterols receptor LXR-alpha, ... | | Authors: | Parthasarathy, G, Klein, D. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Identification and in Vivo Evaluation of Liver X Receptor beta-Selective Agonists for the Potential Treatment of Alzheimer's Disease.
J.Med.Chem., 59, 2016
|
|
6XKC
 
 | | Crystal structure of E3 ligase | | Descriptor: | Protein fem-1 homolog C | | Authors: | Yan, X, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Dong, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for ubiquitin ligase CRL2 FEM1C -mediated recognition of C-degron.
Nat.Chem.Biol., 17, 2021
|
|
5HJP
 
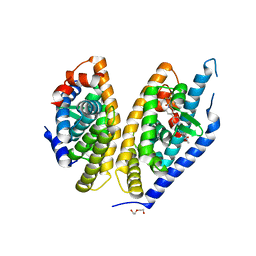 | | Identification of LXRbeta selective agonists for the treatment of Alzheimer's Disease | | Descriptor: | 2-chloro-4-{1'-[(2R)-2-hydroxy-3-methyl-2-(trifluoromethyl)butanoyl]-4,4'-bipiperidin-1-yl}-N,N-dimethylbenzamide, DI(HYDROXYETHYL)ETHER, Oxysterols receptor LXR-beta, ... | | Authors: | Parthasarathy, G, Klein, D. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification and in Vivo Evaluation of Liver X Receptor beta-Selective Agonists for the Potential Treatment of Alzheimer's Disease.
J.Med.Chem., 59, 2016
|
|
5W0X
 
 | | Crystal structure of mouse TOR signaling pathway regulator-like (TIPRL) delta 94-103 | | Descriptor: | TIP41-like protein | | Authors: | Wu, C, Zheng, A, Li, J, Satyshur, K, Xing, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-17 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | Methylation-regulated decommissioning of multimeric PP2A complexes.
Nat Commun, 8, 2017
|
|
4OGV
 
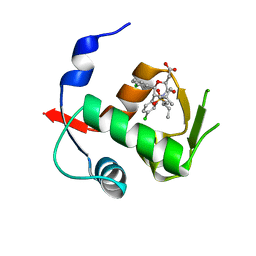 | | Co-Crystal Structure of MDM2 with Inhibitor Compound 49 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, [(2S,5R,6R)-4-[(2S)-1-(tert-butylsulfonyl)butan-2-yl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-3-oxomorpholin-2-yl]acetic acid | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
4OGN
 
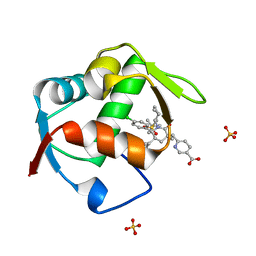 | | Co-Crystal Structure of MDM2 with Inhbitor Compound 3 | | Descriptor: | 6-{[(3R,5R,6S)-1-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]methyl}pyridine-3-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
5HPS
 
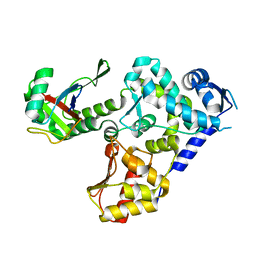 | |
4OGT
 
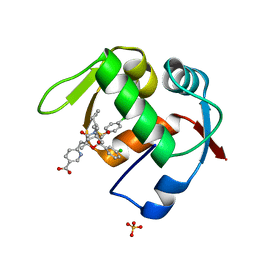 | | Co-Crystal Structure of MDM2 with Inhbitor Compound 46 | | Descriptor: | 6-{[(2R,5R,6R)-4-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-2-methyl-3-oxomorpholin-2-yl]methyl}pyridine-3-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5361 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
5HZG
 
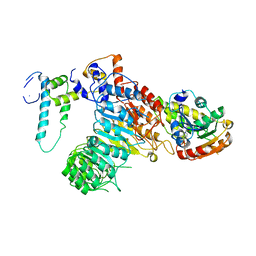 | | The crystal structure of the strigolactone-induced AtD14-D3-ASK1 complex | | Descriptor: | (2Z)-2-methylbut-2-ene-1,4-diol, F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A, ... | | Authors: | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | Deposit date: | 2016-02-02 | | Release date: | 2016-08-03 | | Last modified: | 2016-08-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
4OBA
 
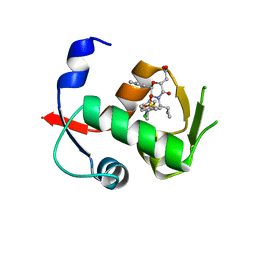 | | Co-crystal structure of MDM2 with Inhibitor Compound 4 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, [(2R,5R,6R)-4-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-3-oxomorpholin-2-yl]acetic acid | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Selective and Potent Morpholinone Inhibitors of the MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 57, 2014
|
|
4OCC
 
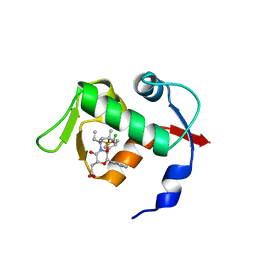 | | co-crystal structure of MDM2(17-111) in complex with compound 48 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, [(2R,5R,6R)-4-[(2S)-1-(tert-butylsulfonyl)butan-2-yl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-3-oxomorpholin-2-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2014-01-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
4ODF
 
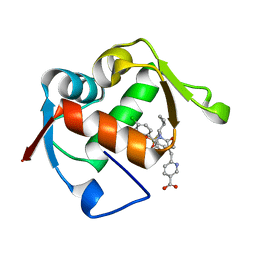 | | Co-Crystal Structure of MDM2 with Inhibitor Compound 47 | | Descriptor: | 6-{[(2S,5R,6R)-4-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-2-methyl-3-oxomorpholin-2-yl]methyl}pyridine-3-carboxylic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2006 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
4HXP
 
 | | Brd4 Bromodomain 1 complex with 4-(2-OXO-1,3-OXAZOLIDIN-3-YL)BENZAMIDE inhibitor | | Descriptor: | 4-(2-oxo-1,3-oxazolidin-3-yl)benzamide, Bromodomain-containing protein 4 | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4HXO
 
 | | Brd4 Bromodomain 1 complex with 3-{[(3-METHYL-1,2-OXAZOL-5-YL)METHYL]SULFANYL}[1,2,4]TRIAZOLO[4,3-A]PYRIDINE inhibitor | | Descriptor: | 3-{[(3-methyl-1,2-oxazol-5-yl)methyl]sulfanyl}[1,2,4]triazolo[4,3-a]pyridine, Bromodomain-containing protein 4 | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
7E7Y
 
 | |
7E7X
 
 | |
7E88
 
 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E86
 
 | |
5I86
 
 | | Crystal structure of the bromodomain of human CREBBP bound to the benzodiazepinone G02778174 | | Descriptor: | (4R)-N-benzyl-4-methyl-2-oxo-2,3,4,5-tetrahydro-1H-1,5-benzodiazepine-6-carboxamide, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
8JP0
 
 | | structure of human sodium-calciumexchanger NCX1 | | Descriptor: | 2-{4-[(2,5-difluorophenyl)methoxy]phenoxy}-5-ethoxyaniline, Sodium/calcium exchanger 1 | | Authors: | Dong, Y, Zhao, Y. | | Deposit date: | 2023-06-09 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insight into the allosteric inhibition of human sodium-calcium exchanger NCX1 by XIP and SEA0400.
Embo J., 43, 2024
|
|
8J4A
 
 | | Crystal structure of OY phytoplasma SAP05 in complex with AtRPN10 | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
