5AX1
 
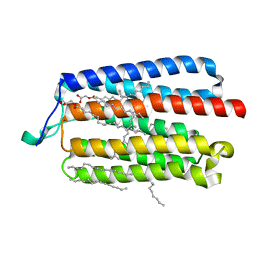 | | Crystal Structure of the Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, at 1.80 angstrom | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DECANE, DODECANE, ... | | Authors: | Furuse, M, Hosaka, T, Kimura-Someya, T, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6PUA
 
 | | The 2.0 A Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Stam, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional characterization of three Type B and C chloramphenicol acetyltransferases from Vibrio species.
Protein Sci., 29, 2020
|
|
7ELJ
 
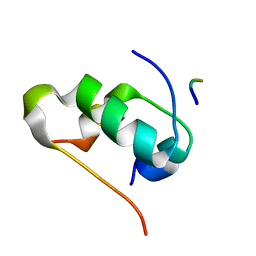 | |
6WI5
 
 | |
3FWW
 
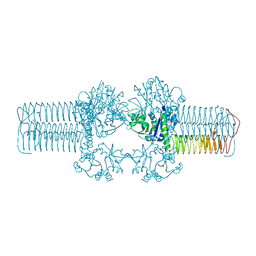 | | The crystal structure of the bifunctional N-acetylglucosamine-1-phosphate uridyltransferase/glucosamine-1-phosphate acetyltransferase from Yersinia pestis CO92 | | Descriptor: | Bifunctional protein glmU | | Authors: | Zhang, R, Gu, M, Stam, J, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-19 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the bifunctional N-acetylglucosamine-1-phosphate uridyltransferase/glucosamine-1-phosphate acetyltransferase from Yersinia pestis CO92
To be Published
|
|
5DMJ
 
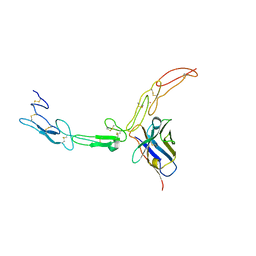 | |
5DMI
 
 | |
5DGD
 
 | |
5DEI
 
 | |
5DGT
 
 | | BENZOYLFORMATE DECARBOXYLASE H70A MUTANT at pH 8.5 FROM PSEUDOMONAS PUTIDA | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Bera, A.K, Hasson, M.S. | | Deposit date: | 2015-08-28 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.081 Å) | | Cite: | BENZOYLFORMATE DECARBOXYLASE H70A MUTANT at pH 8.5 FROM PSEUDOMONAS PUTIDA
to be published
|
|
3AM6
 
 | |
1H70
 
 | | DDAH FROM PSEUDOMONAS AERUGINOSA. C249S MUTANT COMPLEXED WITH CITRULLINE | | Descriptor: | CITRULLINE, NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE | | Authors: | Murray-Rust, J, Leiper, J, McAlister, M, Phelan, J, Tilley, S, Santamaria, J, Vallance, P, McDonald, N. | | Deposit date: | 2001-06-30 | | Release date: | 2001-08-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the hydrolysis of cellular nitric oxide synthase inhibitors by dimethylarginine dimethylaminohydrolase.
Nat. Struct. Biol., 8, 2001
|
|
4JNX
 
 | |
5IHL
 
 | |
5IT2
 
 | | Structure of a transglutaminase 2-specific autoantibody 693-10-B06 Fab fragment | | Descriptor: | heavy chain, light chain | | Authors: | Chen, X, Dalhus, B, Hnida, K, Iversen, R, Sollid, L.M. | | Deposit date: | 2016-03-16 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High abundance of plasma cells secreting transglutaminase 2-specific IgA autoantibodies with limited somatic hypermutation in celiac disease intestinal lesions
Nat. Med., 18, 2012
|
|
8OXW
 
 | |
8OXX
 
 | |
8OXY
 
 | |
8OXV
 
 | |
4NU9
 
 | | 2.30 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus with BME-free Cys289 | | Descriptor: | Betaine aldehyde dehydrogenase, SODIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Stam, J, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-03 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
