1PE3
 
 | |
1TWL
 
 | | Inorganic pyrophosphatase from Pyrococcus furiosus Pfu-264096-001 | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Zhou, W, Tempel, W, Liu, Z.-J, Chen, L, Clancy Kelley, L.-L, Dillard, B.D, Hopkins, R.C, Arendall III, W.B, Rose, J.P, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-07-01 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Inorganic pyrophosphatase from Pyrococcus furiosus Pfu-264096-001
To be published
|
|
2PIS
 
 | | Efforts toward Expansion of the Genetic Alphabet: Structure and Replication of Unnatural Base Pairs | | Descriptor: | DNA (5'-D(*CP*GP*(CBR)P*GP*AP*AP*(FFD)P*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Matsuda, S, Fillo, J.D, Henry, A.A, Wilkins, S.J, Rai, P, Dwyer, T.J, Geierstanger, B.H, Wemmer, D.E, Schultz, P.G, Spraggon, G, Romesberg, F.E. | | Deposit date: | 2007-04-13 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Efforts toward expansion of the genetic alphabet: structure and replication of unnatural base pairs.
J.Am.Chem.Soc., 129, 2007
|
|
2NW2
 
 | | Crystal structure of ELS4 TCR at 1.4A | | Descriptor: | ELS4 TCR alpha chain, ELS4 TCR beta chain | | Authors: | Tynan, F.E, Reid, H.H, Rossjohn, J. | | Deposit date: | 2006-11-14 | | Release date: | 2007-02-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule
Nat.Immunol., 8, 2007
|
|
2R0N
 
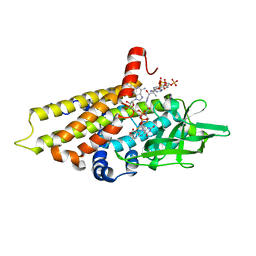 | | The effect of a Glu370Asp mutation in Glutaryl-CoA Dehydrogenase on Proton Transfer to the Dienolate Intermediate | | Descriptor: | 3-thiaglutaryl-CoA, FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase | | Authors: | Rao, K.S, Albro, M, Fu, Z, Narayanan, B, Baddam, S, Lee, H.J, Kim, J.J, Frerman, F.E. | | Deposit date: | 2007-08-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The effect of a Glu370Asp mutation in glutaryl-CoA dehydrogenase on proton transfer to the dienolate intermediate.
Biochemistry, 46, 2007
|
|
2NX5
 
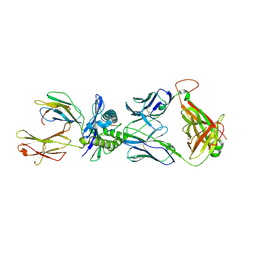 | | Crystal structure of ELS4 TCR bound to HLA-B*3501 presenting EBV peptide EPLPQGQLTAY at 1.7A | | Descriptor: | Beta-2-microglobulin, EBV peptide, EPLPQGQLTAY, ... | | Authors: | Tynan, F.E, Reid, H.H, Rossjohn, J. | | Deposit date: | 2006-11-16 | | Release date: | 2007-02-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule
Nat.Immunol., 8, 2007
|
|
2NW3
 
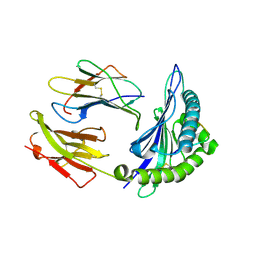 | | Crystal structure of HLA-B*3508 presenting EBV peptide EPLPQGQLTAY at 1.7A | | Descriptor: | Beta-2-microglobulin, EBV peptide EPLPQGQLTAY, HLA class I histocompatibility antigen, ... | | Authors: | Tynan, F.E, Reid, H.H, Rossjohn, J. | | Deposit date: | 2006-11-14 | | Release date: | 2007-02-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule
Nat.Immunol., 8, 2007
|
|
2R0M
 
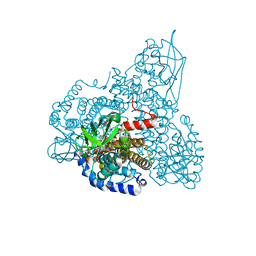 | | The effect of a Glu370Asp Mutation in Glutaryl-CoA Dehydrogenase on Proton Transfer to the Dienolate Intermediate | | Descriptor: | 4-nitrobutanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase | | Authors: | Rao, K.S, Fu, Z, Albro, M, Narayanan, B, Baddam, S, Lee, H.J, Kim, J.J, Frerman, F.E. | | Deposit date: | 2007-08-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The effect of a Glu370Asp mutation in glutaryl-CoA dehydrogenase on proton transfer to the dienolate intermediate.
Biochemistry, 46, 2007
|
|
3ZHN
 
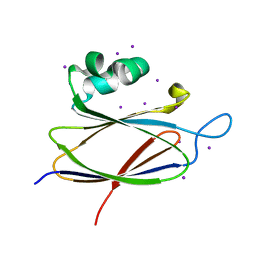 | | Crystal structure of the T6SS lipoprotein TssJ1 from Pseudomonas aeruginosa | | Descriptor: | IODIDE ION, PA_0080 | | Authors: | Robb, C.S, Assmus, M, Nano, F.E, Boraston, A.B. | | Deposit date: | 2012-12-22 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the T6Ss Lipoprotein Tssj1 from Pseudomonas Aeruginosa.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4ACK
 
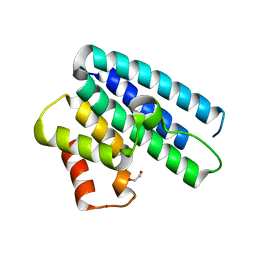 | | 3D Structure of DotU from Francisella novicida | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, TSSL | | Authors: | Robb, C.S, Nano, F.E, Boraston, A.B. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of the Conserved Type Six Secretion Protein Tssl (Dotu) from Francisella Novicida
J.Mol.Biol., 419, 2012
|
|
4YBK
 
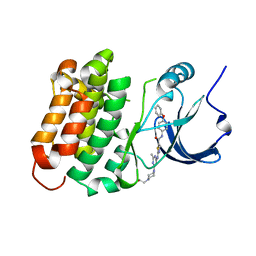 | | C-Helix-Out Dasatinib Analog Crystallized with c-Src Kinase | | Descriptor: | 2-({6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-methylpyrimidin-4-yl}amino)-N-(4-phenoxyphenyl)-1,3-thiazole-5-carboxamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kwarcinski, F.E, Brandvold, K.B, Johnson, T.K, Phadke, S, Meagher, J.L, Seeliger, M.A, Stuckey, J.A, Soellner, M.B. | | Deposit date: | 2015-02-18 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformation-Selective Analogues of Dasatinib Reveal Insight into Kinase Inhibitor Binding and Selectivity.
Acs Chem.Biol., 11, 2016
|
|
4B62
 
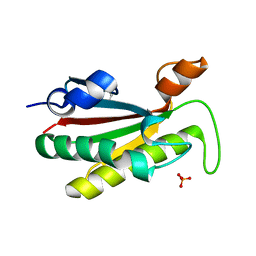 | | The structure of the cell wall anchor of the T6SS from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, TSSL1 | | Authors: | Robb, C.S, Carlson, M, Nano, F.E, Boraston, A.B. | | Deposit date: | 2012-08-08 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of the Periplasmic Peptidoglycan Binding Anchor of a T6Ss from Pseudomonas Aeruginosa.
To be Published
|
|
4YEX
 
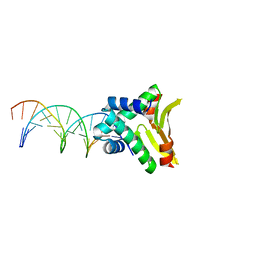 | | HUaa-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YFT
 
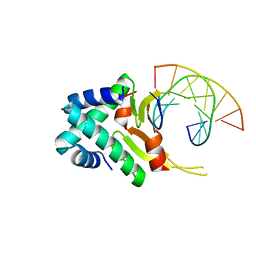 | | HUab-20bp | | Descriptor: | DNA-binding protein HU-alpha, DNA-binding protein HU-beta, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.914 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YEW
 
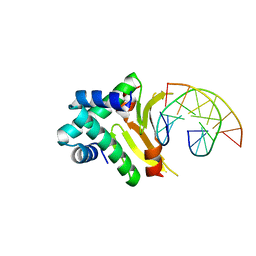 | | HUab-19bp | | Descriptor: | DNA-binding protein HU-alpha, DNA-binding protein HU-beta, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.683 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YC8
 
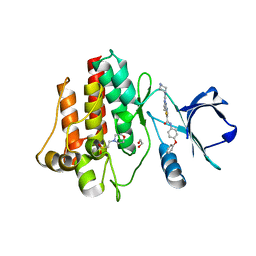 | | C-Helix-Out Binding of Dasatinib Analog to c-Abl Kinase | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-({6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-methylpyrimidin-4-yl}amino)-N-(4-phenoxyphenyl)-1,3-thiazole-5-carboxamide, ... | | Authors: | Kwarcinski, F.E, Brandvold, K.B, Johnson, T.J, Phadke, S, Meagher, J.L, Seeliger, M.A, Stuckey, J.A, Soellner, M.B. | | Deposit date: | 2015-02-19 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conformation-Selective Analogues of Dasatinib Reveal Insight into Kinase Inhibitor Binding and Selectivity.
Acs Chem.Biol., 11, 2016
|
|
4YEY
 
 | | HUaa-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.354 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YFH
 
 | | HU38-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
3HRH
 
 | | Crystal Structure of Antigen 85C and Glycerol | | Descriptor: | Antigen 85-C, GLYCEROL | | Authors: | Boucau, J, Sanki, A.K, Umesiri, F.E, Sucheck, S.J, Ronning, D.R. | | Deposit date: | 2009-06-09 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, synthesis and biological evaluation of sugar-derived esters, alpha-ketoesters and alpha-ketoamides as inhibitors for Mycobacterium tuberculosis antigen 85C.
Mol Biosyst, 5, 2009
|
|
4YF0
 
 | | HU38-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4C8M
 
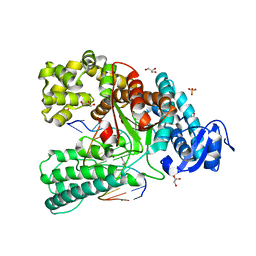 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the aritificial base pair d5SICS-dNaM at the postinsertion site (sequence context 2) | | Descriptor: | GLYCEROL, LARGE FRAGMENT OF TAQ DNA POLYMERASE I, MAGNESIUM ION, ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.568 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4ACL
 
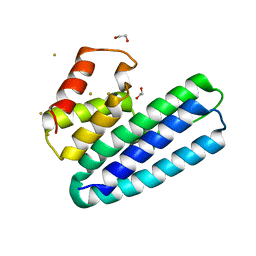 | | 3D Structure of DotU from Francisella novicida | | Descriptor: | 1,2-ETHANEDIOL, GOLD ION, SODIUM ION, ... | | Authors: | Robb, C.S, Nano, F.E, Boraston, A.B. | | Deposit date: | 2011-12-16 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The Structure of the Conserved Type Six Secretion Protein Tssl (Dotu) from Francisella Novicida
J.Mol.Biol., 419, 2012
|
|
3GOT
 
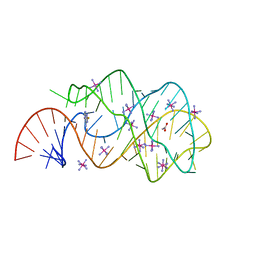 | | Guanine riboswitch C74U mutant bound to 2-fluoroadenine. | | Descriptor: | 2-fluoroadenine, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Gilbert, S.D, Reyes, F.E, Batey, R.T. | | Deposit date: | 2009-03-20 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Adaptive ligand binding by the purine riboswitch in the recognition of Guanine and adenine analogs
Structure, 17, 2009
|
|
4YBJ
 
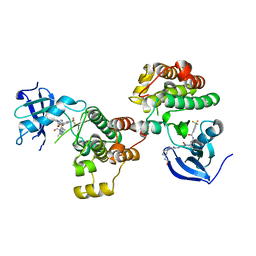 | | Type II Dasatinib Analog Crystallized with c-Src Kinase | | Descriptor: | 2-({6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-methylpyrimidin-4-yl}amino)-N-(3-{[3-(trifluoromethyl)benzoyl]amino}phenyl)-1,3-thiazole-5-carboxamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kwarcinski, F.E, Brandvold, K.B, Johnson, T.K, Phadke, S, Meagher, J.L, Seeliger, M.A, Stuckey, J.A, Soellner, M.B. | | Deposit date: | 2015-02-18 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Conformation-Selective Analogues of Dasatinib Reveal Insight into Kinase Inhibitor Binding and Selectivity.
Acs Chem.Biol., 11, 2016
|
|
3GQX
 
 | |
