7U04
 
 | | IOMA class antibody ACS101 | | Descriptor: | GLYCEROL, IOMA class antibody ACS101 heavy chain, IOMA class antibody ACS101 light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
1AXT
 
 | |
7TI6
 
 | |
1AHW
 
 | | A COMPLEX OF EXTRACELLULAR DOMAIN OF TISSUE FACTOR WITH AN INHIBITORY FAB (5G9) | | Descriptor: | IMMUNOGLOBULIN FAB 5G9 (HEAVY CHAIN), IMMUNOGLOBULIN FAB 5G9 (LIGHT CHAIN), TISSUE FACTOR | | Authors: | Huang, M, Syed, R, Stura, E.A, Stone, M.J, Stefanko, R.S, Ruf, W, Edgington, T.S, Wilson, I.A. | | Deposit date: | 1997-04-10 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The mechanism of an inhibitory antibody on TF-initiated blood coagulation revealed by the crystal structures of human tissue factor, Fab 5G9 and TF.5G9 complex.
J.Mol.Biol., 275, 1998
|
|
1ACY
 
 | |
7JN5
 
 | | Crystal structure of SARS-CoV receptor binding domain in complex with human antibody CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 heavy chain, ... | | Authors: | Wu, N.C, Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | A natural mutation between SARS-CoV-2 and SARS-CoV determines neutralization by a cross-reactive antibody.
Plos Pathog., 16, 2020
|
|
7JMO
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody COVA2-04 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA2-04 heavy chain, COVA2-04 light chain, ... | | Authors: | Wu, N.C, Yuan, M, Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-08-02 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain.
Cell Rep, 33, 2020
|
|
7T7B
 
 | |
7U5G
 
 | | ACS122 Fab | | Descriptor: | ACS122 Fab Heavy chain, ACS122 Fab Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-03-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
7JMP
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody COVA2-39 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA2-39 heavy chain, COVA2-39 light chain, ... | | Authors: | Wu, N.C, Yuan, M, Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-08-02 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain.
Cell Rep, 33, 2020
|
|
7JTF
 
 | |
7JTG
 
 | |
7JPH
 
 | | Crystal structure of EBOV glycoprotein with modified HR1c and HR2 stalk at 3.2 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, ... | | Authors: | Chaudhary, A, Stanfield, R.L, Wilson, I.A, Zhu, J. | | Deposit date: | 2020-08-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.195 Å) | | Cite: | Single-component multilayered self-assembling nanoparticles presenting rationally designed glycoprotein trimers as Ebola virus vaccines.
Nat Commun, 12, 2021
|
|
7JPI
 
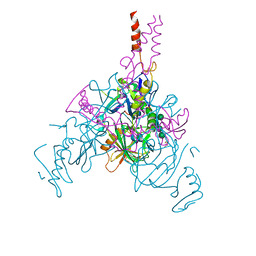 | | Crystal structure of EBOV glycoprotein with modified HR2 stalk at 2.3A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, ... | | Authors: | Chaudhary, A, Stanfield, R.L, Wilson, I.A, Zhu, J. | | Deposit date: | 2020-08-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Single-component multilayered self-assembling nanoparticles presenting rationally designed glycoprotein trimers as Ebola virus vaccines.
Nat Commun, 12, 2021
|
|
7K75
 
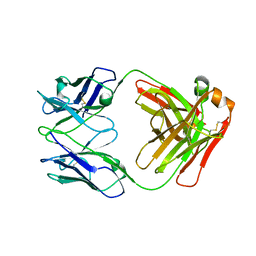 | |
7K76
 
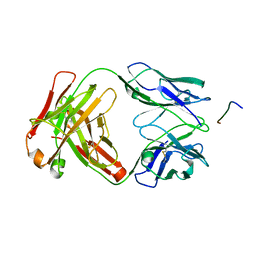 | |
3GAR
 
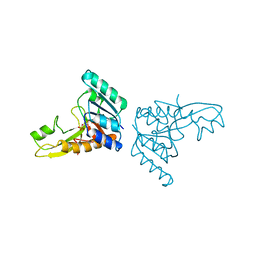 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
7KN5
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain complexed with nanobodies VHH E and U | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
7KN6
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain complexed with nanobody VHH V and antibody Fab CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 Fab heavy chain, CC12.3 Fab light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
7KN7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain complexed with nanobody VHH W and antibody Fab CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 Fab heavy chain, CC12.3 Fab light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
6W00
 
 | | Crystal structure of Fab239 in complex with NPNA2 peptide from circumsporozoite protein | | Descriptor: | Fab239 heavy chain, Fab239 light chain, Immunoglobulin G-binding protein G, ... | | Authors: | Pholcharee, T, Oyen, D, Wilson, I.A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Structural and biophysical correlation of anti-NANP antibodies with in vivo protection against P. falciparum.
Nat Commun, 12, 2021
|
|
6WFX
 
 | |
6WG0
 
 | |
6WFW
 
 | | Crystal structure of Fab364 in complex with NPNA2 peptide from circumsporozoite protein | | Descriptor: | Fab364 heavy chain, Fab364 light chain, Immunoglobulin G-binding protein G, ... | | Authors: | Pholcharee, T, Oyen, D, Wilson, I.A. | | Deposit date: | 2020-04-04 | | Release date: | 2020-07-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Structural and biophysical correlation of anti-NANP antibodies with in vivo protection against P. falciparum.
Nat Commun, 12, 2021
|
|
6WG2
 
 | |
