5D4T
 
 | |
5D4V
 
 | | HcgC with SAH and a guanylylpyridinol (GP) derivative | | Descriptor: | 5'-O-[(R)-[(3,6-dimethyl-2-oxo-1,2-dihydropyridin-4-yl)oxy](hydroxy)phosphoryl]guanosine, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-08-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of HcgC as a SAM-Dependent Pyridinol Methyltransferase in [Fe]-Hydrogenase Cofactor Biosynthesis.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5D5T
 
 | |
5D4U
 
 | | SAM-bound HcgC from Methanocaldococcus jannaschii | | Descriptor: | S-ADENOSYLMETHIONINE, SULFATE ION, Uncharacterized protein MJ0489 | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-08-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of HcgC as a SAM-Dependent Pyridinol Methyltransferase in [Fe]-Hydrogenase Cofactor Biosynthesis.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5D5O
 
 | | HcgC from Methanocaldococcus jannaschii | | Descriptor: | SULFATE ION, Uncharacterized protein MJ0489 | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-08-11 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of HcgC as a SAM-Dependent Pyridinol Methyltransferase in [Fe]-Hydrogenase Cofactor Biosynthesis.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
9BIG
 
 | | Stat6 bound to degrader AK-1690 | | Descriptor: | Signal transducer and activator of transcription 6, [(2-{[(2S)-1-{(2S,4S)-4-[(7-{2-[(3R)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}hept-6-yn-1-yl)oxy]-2-[(2R)-2-phenylmorpholine-4-carbonyl]pyrrolidin-1-yl}-3,3-dimethyl-1-oxobutan-2-yl]carbamoyl}-1-benzothiophen-5-yl)di(fluoro)methyl]phosphonic acid | | Authors: | Mallik, L, Stuckey, J.A. | | Deposit date: | 2024-04-23 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3.304 Å) | | Cite: | Discovery of AK-1690: A Potent and Highly Selective STAT6 PROTAC Degrader.
J.Med.Chem., 2024
|
|
8Y33
 
 | | A near-infrared fluorescent protein of de novo backbone design | | Descriptor: | 3-[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-2-[[5-[(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, near-infrared fluorescent protein | | Authors: | Hu, X, Xu, Y. | | Deposit date: | 2024-01-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Using Protein Design and Directed Evolution to Monomerize a Bright Near-Infrared Fluorescent Protein.
Acs Synth Biol, 13, 2024
|
|
6KXG
 
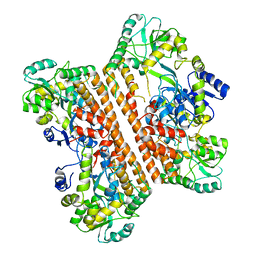 | | Crystal structure of caspase-11-CARD | | Descriptor: | caspase-11-CARD | | Authors: | Liu, M.Z.Y, Jin, T.C. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Crystal structure of caspase-11 CARD provides insights into caspase-11 activation.
Cell Discov, 6, 2020
|
|
6L3M
 
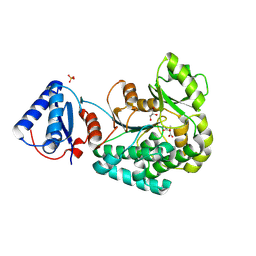 | |
6L3N
 
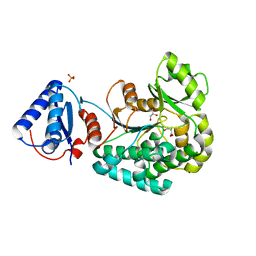 | |
5TCA
 
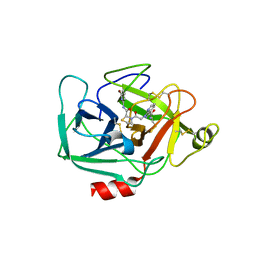 | | Complement Factor D inhibited with JH3 | | Descriptor: | 1-(2-{(2S)-2-[(6-bromopyridin-2-yl)carbamoyl]-1,3-thiazolidin-3-yl}-2-oxoethyl)-1H-pyrazolo[3,4-b]pyridine-3-carboxamide, Complement factor D | | Authors: | Stuckey, J.A. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Buried Hydrogen Bond Interactions Contribute to the High Potency of Complement Factor D Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
5TCC
 
 | | Complement Factor D inhibited with JH4 | | Descriptor: | (2S)-N-(6-bromopyridin-2-yl)-3-[(1H-indazol-1-yl)acetyl]-1,3-thiazolidine-2-carboxamide, Complement factor D | | Authors: | Stuckey, J.A. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Buried Hydrogen Bond Interactions Contribute to the High Potency of Complement Factor D Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
5U8T
 
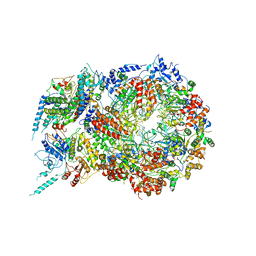 | | Structure of Eukaryotic CMG Helicase at a Replication Fork and Implications | | Descriptor: | Cell division control protein 45, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA replication complex GINS protein PSF1, ... | | Authors: | Li, B, Georgescu, R, Yuan, Z, Santos, R, Sun, J, Zhang, D, Yurieva, O, Li, H, O'Donnell, M.E. | | Deposit date: | 2016-12-15 | | Release date: | 2017-02-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U8S
 
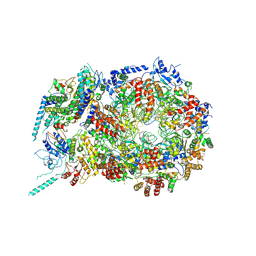 | | Structure of eukaryotic CMG helicase at a replication fork | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA (26-MER), ... | | Authors: | Li, H, Li, B, Georgescu, R, Yuan, Z, Santos, R, Sun, J, Zhang, D, Yurieva, O, O'Donnell, M.E. | | Deposit date: | 2016-12-14 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3SQO
 
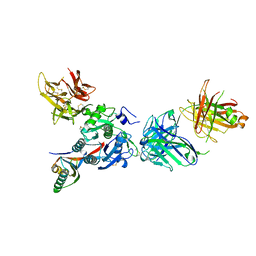 | | PCSK9 J16 Fab complex | | Descriptor: | J16 Heavy chain, J16 Light chain, PCSK9 prodomain, ... | | Authors: | Strop, P. | | Deposit date: | 2011-07-05 | | Release date: | 2012-05-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Proprotein convertase substilisin/kexin type 9 antagonism reduces low-density lipoprotein cholesterol in statin-treated hypercholesterolemic nonhuman primates.
J.Pharmacol.Exp.Ther., 340, 2012
|
|
7SSA
 
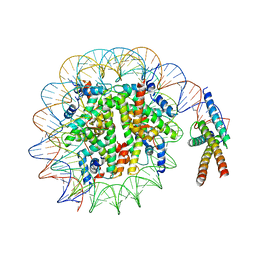 | | Cryo-EM structure of pioneer factor Cbf1 bound to the nucleosome | | Descriptor: | Centromere-binding protein 1, DNA (137-MER), Histone H2A.1, ... | | Authors: | Eek, P, Tan, S. | | Deposit date: | 2021-11-10 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Basic helix-loop-helix pioneer factors interact with the histone octamer to invade nucleosomes and generate nucleosome-depleted regions.
Mol.Cell, 83, 2023
|
|
7UC6
 
 | | Stat5a Core in complex with Compound 12 | | Descriptor: | N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N~3~-(4-bromophenyl)-N,N-dimethyl-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-03-16 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Discovery of a Potent and Selective STAT5 PROTAC Degrader with Strong Antitumor Activity In Vivo in Acute Myeloid Leukemia.
J.Med.Chem., 66, 2023
|
|
7UBT
 
 | | Stat5a Core in complex with Compound 18 | | Descriptor: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N~3~-(1,3-benzothiazol-5-yl)-N,N-dimethyl-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-03-15 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Discovery of a Potent and Selective STAT5 PROTAC Degrader with Strong Antitumor Activity In Vivo in Acute Myeloid Leukemia.
J.Med.Chem., 66, 2023
|
|
7UC7
 
 | | Stat5a Core in complex with Compound 17 | | Descriptor: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N~3~-(1-benzofuran-5-yl)-N,N-dimethyl-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-03-16 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Discovery of a Potent and Selective STAT5 PROTAC Degrader with Strong Antitumor Activity In Vivo in Acute Myeloid Leukemia.
J.Med.Chem., 66, 2023
|
|
7VT1
 
 | |
7VRS
 
 | | The complex of Acyltransferase and Acyl Carrier Protein Domains from module 9 of Salinomycin Polyketide Synthase | | Descriptor: | 1,1'-butane-1,4-diylbis(1H-pyrrole-2,5-dione), 4'-PHOSPHOPANTETHEINE, Type I modular polyketide synthase | | Authors: | Feng, Y, Zheng, J. | | Deposit date: | 2021-10-24 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural visualization of transient interactions between the cis-acting acyltransferase and acyl carrier protein of the salinomycin modular polyketide synthase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8Z4F
 
 | |
7WD0
 
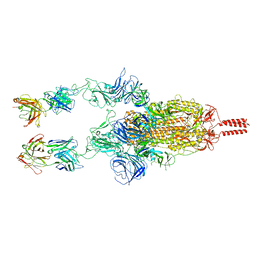 | | SARS-CoV-2 Beta spike in complex with two S5D2 Fabs | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WDF
 
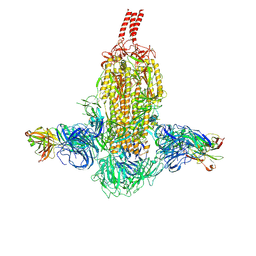 | | SARS-CoV-2 Beta spike in complex with two S3H3 Fabs | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD9
 
 | | SARS-CoV-2 Beta spike in complex with three S3H3 Fabs | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
