8RK7
 
 | |
8RK3
 
 | |
8RQE
 
 | |
7UKL
 
 | |
8RK5
 
 | |
8RKB
 
 | |
8RK6
 
 | |
8RKX
 
 | |
8RK4
 
 | |
1DLY
 
 | | X-RAY CRYSTAL STRUCTURE OF HEMOGLOBIN FROM THE GREEN UNICELLULAR ALGA CHLAMYDOMONAS EUGAMETOS | | Descriptor: | 1,2-ETHANEDIOL, CYANIDE ION, HEMOGLOBIN, ... | | Authors: | Pesce, A, Couture, M, Guertin, M, Dewilde, S, Moens, L, Bolognesi, M. | | Deposit date: | 1999-12-13 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel two-over-two alpha-helical sandwich fold is characteristic of the truncated hemoglobin family.
EMBO J., 19, 2000
|
|
8RKC
 
 | |
8SJ3
 
 | |
8STL
 
 | | Crystal Structure of Nanobody PIK3_Nb16 against wild-type PI3Kalpha | | Descriptor: | Nanobody PIK3_Nb16, SULFATE ION | | Authors: | Nwafor, J.N, Srinivasan, L, Chen, Z, Gabelli, S.B, Iheanacho, A, Alzogaray, V, Klinke, S. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of isoform specific nanobodies for Class I PI3Ks
To be published
|
|
8SLE
 
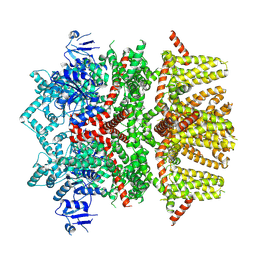 | | Cryo-EM structure of the rat TRPM5 channel in trace calcium, trace-3 | | Descriptor: | Transient receptor potential cation channel subfamily M member 5 | | Authors: | Karuppan, S, Schrag, L.G, Jara-Oseguera, A, Zubcevic, L. | | Deposit date: | 2023-04-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural dynamics at cytosolic interprotomer interfaces control gating of a mammalian TRPM5 channel.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SLI
 
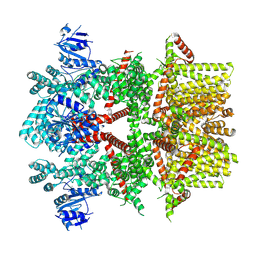 | | Cryo-EM structure of the rat TRPM5 channel in 2mM calcium, high-1 | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily M member 5 | | Authors: | Karuppan, S, Schrag, L.G, Jara-Oseguera, A, Zubcevic, L. | | Deposit date: | 2023-04-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural dynamics at cytosolic interprotomer interfaces control gating of a mammalian TRPM5 channel.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2I04
 
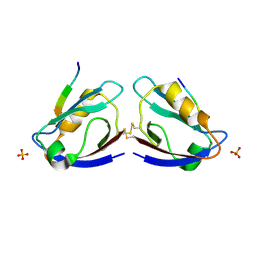 | | X-ray crystal structure of MAGI-1 PDZ1 bound to the C-terminal peptide of HPV18 E6 | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1, SULFATE ION, ... | | Authors: | Chen, X.S, Zhang, Y, Dasgupta, J, Banks, L, Thomas, M. | | Deposit date: | 2006-08-09 | | Release date: | 2007-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of a Human Papillomavirus (HPV) E6 Polypeptide Bound to MAGUK Proteins: Mechanisms of Targeting Tumor Suppressors by a High-Risk HPV Oncoprotein.
J.Virol., 81, 2007
|
|
1DA2
 
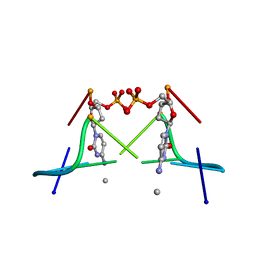 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGMO4CG): N4-METHOXYCYTOSINE/GUANINE BASE-PAIRS IN Z-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(C45)P*G)-3') | | Authors: | Van Meervelt, L, Moore, M.H, Lin, P.K.T, Brown, D.M, Kennard, O. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular and crystal structure of d(CGCGmo4CG): N4-methoxycytosine.guanine base-pairs in Z-DNA.
J.Mol.Biol., 216, 1990
|
|
8SDW
 
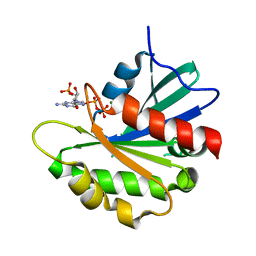 | | Crystal structure of the non-myristoylated mutant [L8K]Arf1 in complex with a GDP analogue | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-3'-MONOPHOSPHATE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Rosenberg Jr, E.M, Randazzo, P.A, Esser, L, Xia, D. | | Deposit date: | 2023-04-07 | | Release date: | 2023-06-28 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Point mutations in Arf1 reveal cooperative effects of the N-terminal extension and myristate for GTPase-activating protein catalytic activity.
Plos One, 19, 2024
|
|
8SLQ
 
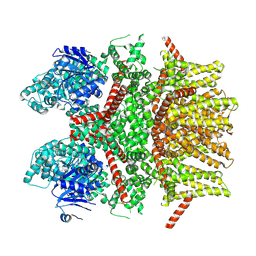 | | Cryo-EM structure of the rat TRPM5 channel in 2mM calcium, high-3 | | Descriptor: | Transient receptor potential cation channel subfamily M member 5 | | Authors: | Karuppan, S, Schrag, L.G, Jara-Oseguera, A, Zubcevic, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural dynamics at cytosolic interprotomer interfaces control gating of a mammalian TRPM5 channel.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1DI8
 
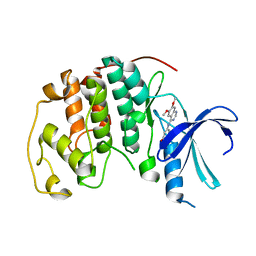 | | THE STRUCTURE OF CYCLIN-DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH 4-[3-HYDROXYANILINO]-6,7-DIMETHOXYQUINAZOLINE | | Descriptor: | 4-[3-HYDROXYANILINO]-6,7-DIMETHOXYQUINAZOLINE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Shewchuk, L, Hassell, A, Kuyper, L.F. | | Deposit date: | 1999-11-29 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding mode of the 4-anilinoquinazoline class of protein kinase inhibitor: X-ray crystallographic studies of 4-anilinoquinazolines bound to cyclin-dependent kinase 2 and p38 kinase.
J.Med.Chem., 43, 2000
|
|
1DK4
 
 | | CRYSTAL STRUCTURE OF MJ0109 GENE PRODUCT INOSITOL MONOPHOSPHATASE | | Descriptor: | INOSITOL MONOPHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Stec, B, Yang, H, Johnson, K.A, Chen, L, Roberts, M.F. | | Deposit date: | 1999-12-06 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MJ0109 is an enzyme that is both an inositol monophosphatase and the 'missing' archaeal fructose-1,6-bisphosphatase.
Nat.Struct.Biol., 7, 2000
|
|
2I0L
 
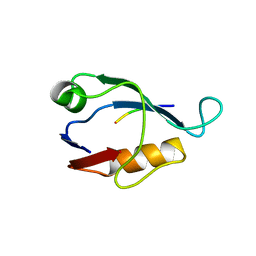 | | X-ray crystal structure of Sap97 PDZ2 bound to the C-terminal peptide of HPV18 E6. | | Descriptor: | Disks large homolog 1, peptide E6 | | Authors: | Chen, X.S, Zhang, Y, Dasgupta, J, Banks, L, Thomas, M. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structures of a Human Papillomavirus (HPV) E6 Polypeptide Bound to MAGUK Proteins: Mechanisms of Targeting Tumor Suppressors by a High-Risk HPV Oncoprotein.
J.Virol., 81, 2007
|
|
1DJ0
 
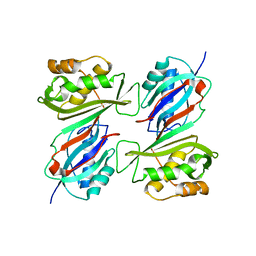 | | THE CRYSTAL STRUCTURE OF E. COLI PSEUDOURIDINE SYNTHASE I AT 1.5 ANGSTROM RESOLUTION | | Descriptor: | CHLORIDE ION, PSEUDOURIDINE SYNTHASE I | | Authors: | Foster, P.G, Huang, L, Santi, D.V, Stroud, R.M. | | Deposit date: | 1999-11-30 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I.
Nat.Struct.Biol., 7, 2000
|
|
1DJP
 
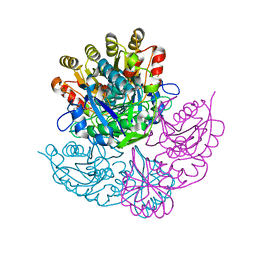 | | CRYSTAL STRUCTURE OF PSEUDOMONAS 7A GLUTAMINASE-ASPARAGINASE WITH THE INHIBITOR DON COVALENTLY BOUND IN THE ACTIVE SITE | | Descriptor: | 5,5-dihydroxy-6-oxo-L-norleucine, GLUTAMINASE-ASPARAGINASE | | Authors: | Ortlund, E, Lacount, M.W, Lewinski, K, Lebioda, L. | | Deposit date: | 1999-12-03 | | Release date: | 2000-01-24 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reactions of Pseudomonas 7A glutaminase-asparaginase with diazo analogues of glutamine and asparagine result in unexpected covalent inhibitions and suggests an unusual catalytic triad Thr-Tyr-Glu.
Biochemistry, 39, 2000
|
|
8SL8
 
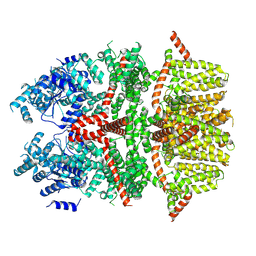 | | Cryo-EM structure of the rat TRPM5 channel in trace calcium, trace-1 | | Descriptor: | Transient receptor potential cation channel subfamily M member 5 | | Authors: | Karuppan, S, Schrag, L.G, Jara-Oseguera, A, Zubcevic, L. | | Deposit date: | 2023-04-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural dynamics at cytosolic interprotomer interfaces control gating of a mammalian TRPM5 channel.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
