1XN9
 
 | | Solution Structure of Methanosarcina mazei Protein RPS24E: The Northeast Structural Genomics Consortium Target MaR11 | | Descriptor: | 30S ribosomal protein S24e | | Authors: | Liu, G, Xiao, R, Parish, D, Ma, L, Sukumaran, D, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Methanosarcina mazei Protein RPS24E: The Northeast Structural Genomics Consortium Target MaR11
To be Published
|
|
1UMO
 
 | | The crystal structure of cytoglobin: the fourth globin type discovered in man | | Descriptor: | CYTOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | de Sanctis, D, Dewilde, S, Pesce, A, Moens, L, Ascenzi, P, Hankeln, T, Burmester, T, Bolognesi, M. | | Deposit date: | 2003-08-26 | | Release date: | 2004-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of cytoglobin: the fourth globin type discovered in man displays heme hexa-coordination.
J.Mol.Biol., 336, 2004
|
|
1XYJ
 
 | | NMR Structure of the cat prion protein | | Descriptor: | prion protein | | Authors: | Lysek, D.A, Schorn, C, Nivon, L.G, Esteve-Moya, V, Christen, B, Calzolai, L, von Schroetter, C, Fiorito, F, Herrmann, T, Guntert, P. | | Deposit date: | 2004-11-10 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5DV9
 
 | |
6BQ9
 
 | | 2.55 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-493) of DNA Topoisomerase IV Subunit A from Pseudomonas putida | | Descriptor: | CHLORIDE ION, DNA topoisomerase 4 subunit A, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 2.55 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-493) of DNA Topoisomerase IV Subunit A from Pseudomonas putida.
To Be Published
|
|
1XO2
 
 | | Crystal structure of a human cyclin-dependent kinase 6 complex with a flavonol inhibitor, fisetin | | Descriptor: | 3,7,3',4'-TETRAHYDROXYFLAVONE, Cell division protein kinase 6, Cyclin | | Authors: | Lu, H.S, Chang, D.J, Baratte, B, Meijer, L, Schulze-Gahmen, U. | | Deposit date: | 2004-10-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Human Cyclin-Dependent Kinase 6 Complex with a Flavonol Inhibitor, Fisetin.
J.Med.Chem., 48, 2005
|
|
6BV4
 
 | | Crystal structure of porcine aminopeptidase-N with Methionine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
1UKV
 
 | | Structure of RabGDP-dissociation inhibitor in complex with prenylated YPT1 GTPase | | Descriptor: | GERAN-8-YL GERAN, GTP-binding protein YPT1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Rak, A, Pylypenko, O, Durek, T, Watzke, A, Kushnir, S, Brunsveld, L, Waldmann, H, Goody, R.S, Alexandrov, K. | | Deposit date: | 2003-09-01 | | Release date: | 2004-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase
Science, 302, 2003
|
|
6BYV
 
 | | Solution NMR structure of cysteine-rich calcium bound domains of very low density lipoprotein receptor | | Descriptor: | CALCIUM ION, Very low-density lipoprotein receptor | | Authors: | Banerjee, K, Gruschus, J.M, Tjandra, N, Yakovlev, S, Medved, L. | | Deposit date: | 2017-12-21 | | Release date: | 2018-07-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of the Recombinant Fragment Containing Three Fibrin-Binding Cysteine-Rich Domains of the Very Low Density Lipoprotein Receptor.
Biochemistry, 57, 2018
|
|
8P4E
 
 | | Structural insights into human co-transcriptional capping - structure 5 | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (26-MER), DNA (35-MER), ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
1XI9
 
 | | Alanine aminotransferase from Pyrococcus furiosus Pfu-1397077-001 | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, putative transaminase | | Authors: | Zhou, W, Tempel, W, Shah, A, Chen, L, Liu, Z.-J, Lee, D, Lin, D, Chang, S.-H, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Lee, H.-S, Poole II, F.L, Shah, C, Sugar, F.J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Alanine aminotransferase from Pyrococcus furiosus Pfu-1397077-001
To be published
|
|
6C0U
 
 | | Crystal structure of cAMP-dependent protein kinase Calpha subunit bound with N46 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3R,4R)-4-{[4-(2-fluoro-3-methoxy-6-propoxybenzene-1-carbonyl)benzene-1-carbonyl]amino}pyrrolidin-3-yl]-1H-indazole-5-carboxamide, PHOSPHATE ION, ... | | Authors: | Qin, L, Sankaran, B, Kim, C. | | Deposit date: | 2018-01-02 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for selective inhibition of human PKG I alpha by the balanol-like compound N46.
J. Biol. Chem., 293, 2018
|
|
1RI8
 
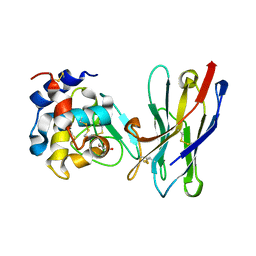 | | Crystal Structure of the Camelid Single Domain Antibody 1D2L19 in complex with Hen Egg White Lysozyme | | Descriptor: | GLYCEROL, Lysozyme C, camelid ANTIBODY HEAVY CHAIN | | Authors: | De Genst, E, Silence, K, Ghahroudi, M.A, Decanniere, K, Loris, R, Kinne, J, Wyns, L, Muyldermans, S. | | Deposit date: | 2003-11-17 | | Release date: | 2005-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Strong in vivo maturation compensates for structurally restricted H3 loops in antibody repertoires.
J.Biol.Chem., 280, 2005
|
|
8QED
 
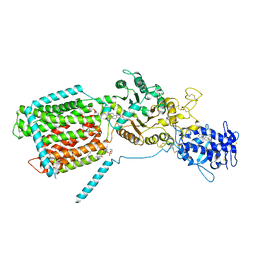 | | S. cerevisia Niemann-Pick type C protein NCR1 in LMNG at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ERGOSTEROL, NPC intracellular sterol transporter 1-related protein 1, ... | | Authors: | Frain, K.M, Nel, L, Dedic, E, Olesen, E, Stokes, D, Panyella Pedersen, B. | | Deposit date: | 2023-08-31 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Conformational changes in the Niemann-Pick type C1 protein NCR1 drive sterol translocation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6C26
 
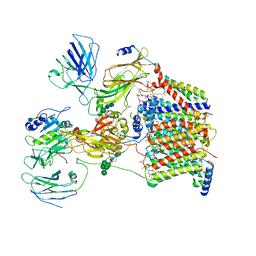 | | The Cryo-EM structure of a eukaryotic oligosaccharyl transferase complex | | Descriptor: | (4R,7R)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(undecanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphadocosan-1-aminium, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2018-01-06 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The atomic structure of a eukaryotic oligosaccharyltransferase complex.
Nature, 555, 2018
|
|
8QEE
 
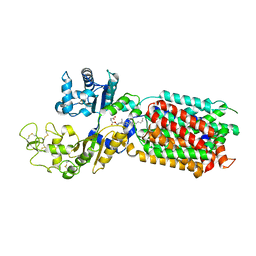 | | S. cerevisia Niemann-Pick type C protein NCR1 in Peptidisc at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Frain, K.M, Dedic, E, Nel, L, Olesen, E, Stokes, D, Panyella Pedersen, B. | | Deposit date: | 2023-08-31 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Conformational changes in the Niemann-Pick type C1 protein NCR1 drive sterol translocation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1XS7
 
 | | Crystal Structure of a cycloamide-urethane-derived novel inhibitor bound to human brain memapsin 2 (beta-secretase). | | Descriptor: | Beta-secretase 1, N-[(4S,5S,7R)-8-({(S)-1-[(BENZYLAMINO)OXOMETHYL]-2-METHYLPROPYL}AMINO)-5-HYDROXY-2,7-DIMETHYL-8-OXO-OCT-4-YL]-(4S,7S)-4 -ISOPROPYL-2,5,9-TRIOXO-1-OXA-3,6,10-TRIAZACYCLOHEXADECANE-7-CARBOXAMIDE | | Authors: | Ghosh, A, Devasamudram, T, Hong, L, DeZutter, C, Xu, X, Weerasena, V, Koelsch, G, Bilcer, G, Tang, J. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of cycloamide-urethane-derived novel inhibitors of human brain memapsin 2 (beta-secretase).
Bioorg.Med.Chem.Lett., 15, 2005
|
|
6C8T
 
 | | The structure of MppP soaked with the substrate L-Arg | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, CHLORIDE ION, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
8Q4H
 
 | | a membrane-bound menaquinol:organohalide oxidoreductase complex RDH complex | | Descriptor: | (~{Z})-1,2-bis(chloranyl)ethene, CO-METHYLCOBALAMIN, IRON/SULFUR CLUSTER, ... | | Authors: | Dongchun, N, Ekundayo, B, Henning, S, Julien, M, Holliger, C, Cimmino, L. | | Deposit date: | 2023-08-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structure of a membrane-bound menaquinol:organohalide oxidoreductase.
Nat Commun, 14, 2023
|
|
1XSE
 
 | | Crystal Structure of Guinea Pig 11beta-Hydroxysteroid Dehydrogenase Type 1 | | Descriptor: | 11beta-hydroxysteroid dehydrogenase type 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ogg, D, Elleby, B, Norstrom, C, Stefansson, K, Abrahmsen, L, Oppermann, U, Svensson, S. | | Deposit date: | 2004-10-19 | | Release date: | 2004-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of guinea pig 11beta-hydroxysteroid dehydrogenase type 1 provides a model for enzyme-lipid bilayer interactions
J.Biol.Chem., 280, 2005
|
|
1RK7
 
 | | Solution structure of apo Cu,Zn Superoxide Dismutase: role of metal ions in protein folding | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Banci, L, Bertini, I, Cramaro, F, Del Conte, R, Viezzoli, M.S. | | Deposit date: | 2003-11-21 | | Release date: | 2003-12-02 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Apo Cu,Zn Superoxide Dismutase: Role of Metal Ions in Protein Folding
Biochemistry, 42, 2003
|
|
8QEC
 
 | | S. cerevisia Niemann-Pick type C protein NCR1 in GDN at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Frain, K.M, Dedic, E, Nel, L, Olesen, E, Stokes, D, Panyella Pedersen, B. | | Deposit date: | 2023-08-31 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational changes in the Niemann-Pick type C1 protein NCR1 drive sterol translocation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1RQH
 
 | | Propionibacterium shermanii transcarboxylase 5S subunit bound to pyruvic acid | | Descriptor: | COBALT (II) ION, PYRUVIC ACID, transcarboxylase 5S subunit | | Authors: | Hall, P.R, Zheng, R, Antony, L, Pusztai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2003-12-05 | | Release date: | 2004-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit.
Embo J., 23, 2004
|
|
1XNW
 
 | | Acyl-CoA Carboxylase Beta Subunit from S. coelicolor (PccB), apo form #2, mutant D422I | | Descriptor: | propionyl-CoA carboxylase complex B subunit | | Authors: | Diacovich, L, Mitchell, D.L, Pham, H, Gago, G, Melgar, M.M, Khosla, C, Gramajo, H, Tsai, S.-C. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the beta-Subunit of Acyl-CoA Carboxylase: Structure-Based Engineering of Substrate Specificity
Biochemistry, 43, 2004
|
|
1XV5
 
 | | alpha-glucosyltransferase (AGT) in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA alpha-glucosyltransferase, ... | | Authors: | Lariviere, L, Sommer, N, Morera, S. | | Deposit date: | 2004-10-27 | | Release date: | 2005-08-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural evidence of a passive base-flipping mechanism for AGT, an unusual GT-B glycosyltransferase.
J.Mol.Biol., 352, 2005
|
|
