5EAM
 
 | | Crystal structure of human WDR5 in complex with compound 9o | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
5EB1
 
 | | the YfiB-YfiR complex | | Descriptor: | SULFATE ION, YfiB, YfiR | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
1VYT
 
 | | beta3 subunit complexed with aid | | Descriptor: | CALCIUM CHANNEL BETA-3 SUBUNIT, VOLTAGE-DEPENDENT L-TYPE CALCIUM CHANNEL ALPHA-1C SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Tong, L, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
1R6A
 
 | | Structure of the dengue virus 2'O methyltransferase in complex with s-adenosyl homocysteine and ribavirin 5' triphosphate | | Descriptor: | Genome polyprotein, RIBAVIRIN MONOPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Benarroch, D, Egloff, M.P, Mulard, L, Romette, J.L, Canard, B. | | Deposit date: | 2003-10-15 | | Release date: | 2004-09-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A structural basis for the inhibition of the NS5 dengue virus mRNA 2'-O-methyltransferase domain by ribavirin 5'-triphosphate.
J.Biol.Chem., 279, 2004
|
|
6DA7
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with apo form at 1.83 A resolution (I222) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
6DUX
 
 | | 2.25 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Klebsiella pneumoniae in Complex with NAD. | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 6-phospho-alpha-glucosidase, ACETATE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-22 | | Release date: | 2018-07-04 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
1R7J
 
 | | Crystal structure of the DNA-binding protein Sso10a from Sulfolobus solfataricus | | Descriptor: | Conserved hypothetical protein Sso10a | | Authors: | Chen, L, Chen, L.R, Zhou, X.E, Wang, Y, Kahsai, M.A, Clark, A.T, Edmondson, S.P, Liu, Z.-J, Rose, J.P, Wang, B.C, Shriver, J.W, Meehan, E.J, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-10-21 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The hyperthermophile protein Sso10a is a dimer of winged helix DNA-binding domains linked by an antiparallel coiled coil rod.
J.Mol.Biol., 341, 2004
|
|
1W31
 
 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE 5-HYDROXYLAEVULINIC ACID COMPLEX | | Descriptor: | 5-HYDROXYLAEVULINIC ACID, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Stauffer, F, Beaven, G.D.E, Gill, R, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2004-07-11 | | Release date: | 2005-08-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with the Inhibitor 5-Hydroxylaevulinic Acid
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6BUY
 
 | | Crystal structure of porcine aminopeptidase-N with Glycine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-11 | | Release date: | 2018-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
2NQY
 
 | |
1W51
 
 | | BACE (Beta Secretase) in complex with a nanomolar non-peptidic inhibitor | | Descriptor: | 3-[({(1S,2R)-1-BENZYL-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}AMINO)(HYDROXY)METHYL]-N,N-DIPROPYLBENZAMIDE, BETA-SECRETASE 1, IODIDE ION | | Authors: | Patel, S, Vuillard, L, Cleasby, A, Murray, C.W, Yon, J. | | Deposit date: | 2004-08-04 | | Release date: | 2004-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Apo and Inhibitor Complex Structures of Bace (Beta-Secretase)
J.Mol.Biol., 343, 2004
|
|
1W2B
 
 | | Trigger Factor ribosome binding domain in complex with 50S | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L13P, ... | | Authors: | Ferbitz, L, Maier, T, Patzelt, H, Bukau, B, Deuerling, E, Ban, N. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Trigger Factor in Complex with the Ribosome Forms a Molecular Cradle for Nascent Proteins
Nature, 431, 2004
|
|
6BWT
 
 | | 2.45 Angstrom Resolution Crystal Structure Thioredoxin Reductase from Francisella tularensis. | | Descriptor: | CHLORIDE ION, SULFATE ION, Thioredoxin reductase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-15 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | 2.45 Angstrom Resolution Crystal Structure Thioredoxin Reductase from Francisella tularensis.
To Be Published
|
|
1S3H
 
 | | Propionibacterium shermanii transcarboxylase 5S subunit A59T | | Descriptor: | COBALT (II) ION, transcarboxylase 5S subunit | | Authors: | Hall, P.R, Zheng, R, Antony, L, Pusztai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2004-01-13 | | Release date: | 2004-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit.
Embo J., 23, 2004
|
|
5D8N
 
 | | Tomato leucine aminopeptidase mutant - K354E | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | DuPrez, K.T, Scranton, M.A, Walling, L.L, Fan, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into chaperone-activity enhancement by a K354E mutation in tomato acidic leucine aminopeptidase.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1S1C
 
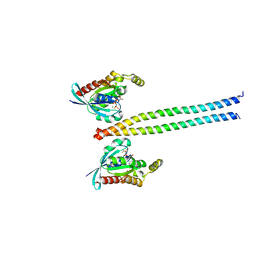 | | Crystal structure of the complex between the human RhoA and Rho-binding domain of human ROCKI | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Rho-associated, ... | | Authors: | Dvorsky, R, Blumenstein, L, Vetter, I.R, Ahmadian, M.R. | | Deposit date: | 2004-01-06 | | Release date: | 2004-02-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into the Interaction of ROCKI with the Switch Regions of RhoA.
J.Biol.Chem., 279, 2004
|
|
8PI9
 
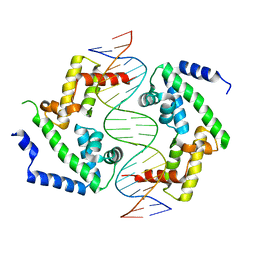 | | DNA binding domain of HNF-1A bound to P2-HNF4A promoter DNA variant (P2 -181G>A) | | Descriptor: | Chains: E, Chains: F, Hepatocyte nuclear factor 1-alpha | | Authors: | Kind, L, Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular mechanism of HNF-1A-mediated HNF4A gene regulation and promoter-driven HNF4A-MODY diabetes.
JCI Insight, 9, 2024
|
|
1S62
 
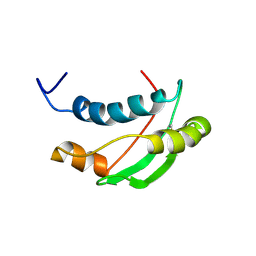 | | Solution structure of the Escherichia coli TolA C-terminal domain | | Descriptor: | TolA protein | | Authors: | Deprez, C, Blanchard, L, Simorre, J.-P, Gavioli, M, Guerlesquin, F, Lazdunski, C, Lloubes, R, Marion, D. | | Deposit date: | 2004-01-22 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the E.coli TolA C-terminal domain reveals conformational changes upon binding to the phage g3p N-terminal domain.
J.Mol.Biol., 346, 2005
|
|
5DFZ
 
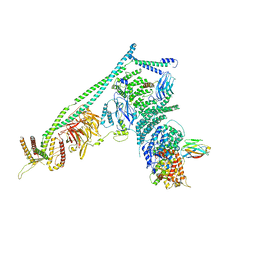 | | Structure of Vps34 complex II from S. cerevisiae. | | Descriptor: | Nanobody binding S. cerevisiae Vps34, Phosphatidylinositol 3-kinase VPS34, Putative N-terminal domain of S. cerevisiae Vps30, ... | | Authors: | Rostislavleva, K, Soler, N, Ohashi, Y, Zhang, L, Williams, R.L. | | Deposit date: | 2015-08-27 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes.
Science, 350, 2015
|
|
6C0F
 
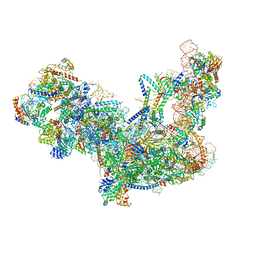 | | Yeast nucleolar pre-60S ribosomal subunit (state 2) | | Descriptor: | 5.8S rRNA, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Sanghai, Z.A, Miller, L, Barandun, J, Hunziker, M, Chaker-Margot, M, Klinge, S. | | Deposit date: | 2017-12-29 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Modular assembly of the nucleolar pre-60S ribosomal subunit.
Nature, 556, 2018
|
|
1VR4
 
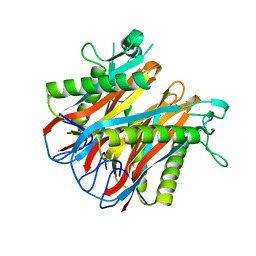 | | Crystal Structure of MCSG TArget APC22750 from Bacillus cereus | | Descriptor: | hypothetical protein APC22750 | | Authors: | Yang, X, Brunzelle, J.S, McNamara, L.K, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-01-31 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of MCSG TArget APC22750 from Bacillus cereus
To be Published
|
|
1S0P
 
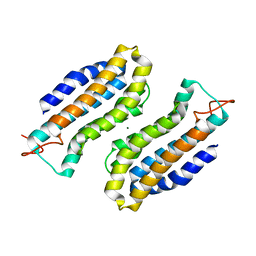 | | Structure of the N-Terminal Domain of the Adenylyl Cyclase-Associated Protein (CAP) from Dictyostelium discoideum. | | Descriptor: | Adenylyl cyclase-associated protein, MAGNESIUM ION | | Authors: | Ksiazek, D, Brandstetter, H, Israel, L, Bourenkov, G.P, Katchalova, G, Janssen, K.P, Bartunik, H.D, Noegel, A.A, Schleicher, M, Holak, T.A. | | Deposit date: | 2004-01-01 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | STRUCTURE OF THE N-TERMINAL DOMAIN OF THE ADENYLYL
CYCLASE-ASSOCIATED PROTEIN (CAP) FROM DICTYOSTELIUM DISCOIDEUM
Structure, 11, 2003
|
|
5DCY
 
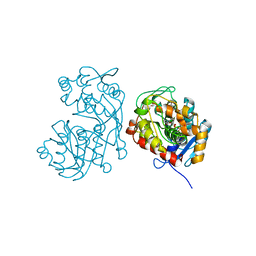 | | Iridoid synthase G150A mutant from Catharanthus roseus - binary complex with NADP+ | | Descriptor: | 1,2-ETHANEDIOL, Iridoid synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Caputi, L, Kries, H, Stevenson, C.E.M, Kamileen, M.O, Sherden, N.H, Geu-Flores, F, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2015-08-24 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural determinants of reductive terpene cyclization in iridoid biosynthesis.
Nat.Chem.Biol., 12, 2016
|
|
5DJ3
 
 | | Structure of the PLP-Dependent L-Arginine Hydroxylase MppP with D-Arginine Bound | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-D-arginine, MAGNESIUM ION, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Silvaggi, N.R, Han, L. | | Deposit date: | 2015-09-01 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Streptomyces wadayamensis MppP Is a Pyridoxal 5'-Phosphate-Dependent l-Arginine alpha-Deaminase, gamma-Hydroxylase in the Enduracididine Biosynthetic Pathway.
Biochemistry, 54, 2015
|
|
6C95
 
 | |
