6B50
 
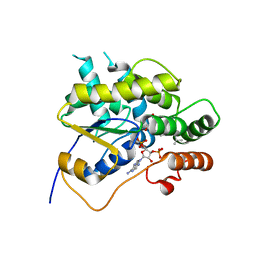 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase/Oxamniquine Complex, T157S Mutant | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase oxamniquine resistance protein, {(2S)-7-nitro-2-[(propan-2-ylamino)methyl]-1,2,3,4-tetrahydroquinolin-6-yl}methanol | | Authors: | Taylor, A.B. | | Deposit date: | 2017-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Why does oxamniquine kill Schistosoma mansoni and not S. haematobium and S. japonicum?
Int.J.Parasitol., 2020
|
|
6BDQ
 
 | |
6B4Z
 
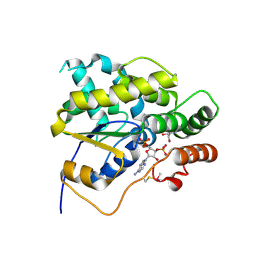 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase, T157S Mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase oxamniquine resistance protein | | Authors: | Taylor, A.B. | | Deposit date: | 2017-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Why does oxamniquine kill Schistosoma mansoni and not S. haematobium and S. japonicum?
Int.J.Parasitol., 2020
|
|
6B54
 
 | |
6BPZ
 
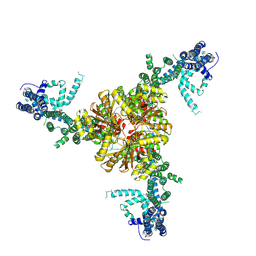 | | Structure of the mechanically activated ion channel Piezo1 | | Descriptor: | Piezo-type mechanosensitive ion channel component 1,Piezo-type mechanosensitive ion channel component 1,mouse Piezo1,Piezo-type mechanosensitive ion channel component 1,Piezo-type mechanosensitive ion channel component 1 | | Authors: | Saotome, K, Kefauver, J.M, Patapoutian, A, Ward, A.B. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the mechanically activated ion channel Piezo1.
Nature, 554, 2018
|
|
6BU8
 
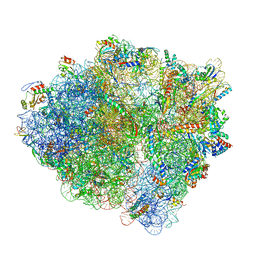 | | 70S ribosome with S1 domains 1 and 2 (Class 1) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Loveland, A.B, Korostelev, A.A. | | Deposit date: | 2017-12-08 | | Release date: | 2018-01-31 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural dynamics of protein S1 on the 70S ribosome visualized by ensemble cryo-EM.
Methods, 137, 2018
|
|
6CJV
 
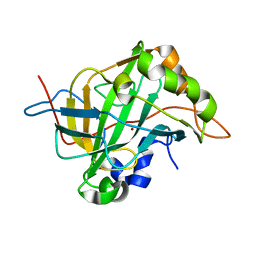 | | Carbonic anhydrase IX-mimic in complex with sucralose | | Descriptor: | 4-chloro-4-deoxy-alpha-D-galactopyranose-(1-2)-1,6-dichloro-1,6-dideoxy-beta-D-fructofuranose, Carbonic anhydrase 2, ZINC ION | | Authors: | Lomelino, C.L, Murray, A.B, McKenna, R. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Sweet Binders: Carbonic Anhydrase IX in Complex with Sucralose.
ACS Med Chem Lett, 9, 2018
|
|
6CRV
 
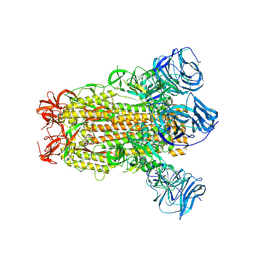 | | SARS Spike Glycoprotein, Stabilized variant, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6C5V
 
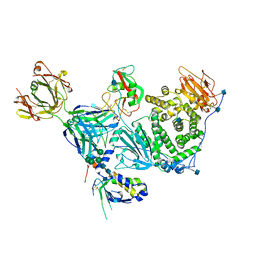 | | An anti-gH/gL antibody that neutralizes dual-tropic infection defines a site of vulnerability on Epstein-Barr virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab AMMO1 heavy chain, Antibody Fab AMMO1 light chain, ... | | Authors: | Snijder, J, Ortego, M.S, Weidle, C, Stuart, A.B, Gray, M.A, McElrath, M.J, Pancera, M, Veesler, D, McGuire, A.T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-01-16 | | Release date: | 2018-05-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | An Antibody Targeting the Fusion Machinery Neutralizes Dual-Tropic Infection and Defines a Site of Vulnerability on Epstein-Barr Virus.
Immunity, 48, 2018
|
|
6CRZ
 
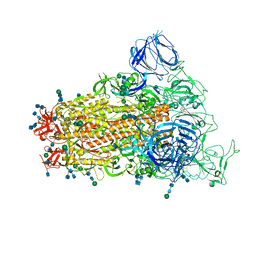 | | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CS0
 
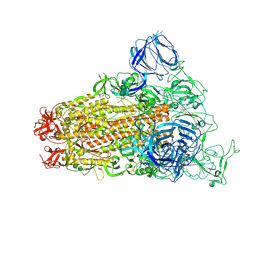 | | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, one S1 CTD in an upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CJK
 
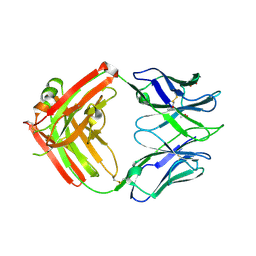 | | Anti HIV Fab 10A | | Descriptor: | ACETATE ION, GLYCEROL, Immunoglobulin Fab heavy chain, ... | | Authors: | Hangartner, L, Ward, A.B, Wilson, I.A, Oyen, D. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-15 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization.
Immunity, 49, 2018
|
|
6CS2
 
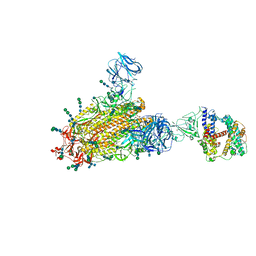 | | SARS Spike Glycoprotein - human ACE2 complex, Stabilized variant, all ACE2-bound particles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6AWB
 
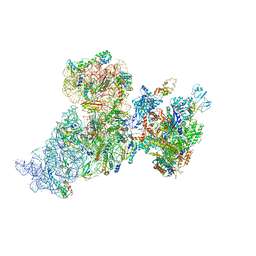 | | Structure of 30S ribosomal subunit and RNA polymerase complex in non-rotated state | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6B52
 
 | |
6APO
 
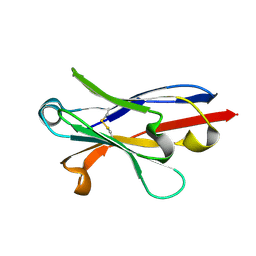 | | Anti-Marburgvirus Nucleoprotein Single Domain Antibody A | | Descriptor: | Anti-Marburgvirus Nucleoprotein Single Domain Antibody A | | Authors: | Taylor, A.B, Garza, J.A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.168 Å) | | Cite: | Unveiling a Drift Resistant Cryptotope withinMarburgvirusNucleoprotein Recognized by Llama Single-Domain Antibodies.
Front Immunol, 8, 2017
|
|
6CRQ
 
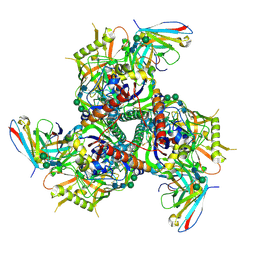 | | Glutaraldehyde-treated BG505 SOSIP.664 Env in complex with PGV04 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Pallesen, J, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-18 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural and immunologic correlates of chemically stabilized HIV-1 envelope glycoproteins.
PLoS Pathog., 14, 2018
|
|
6CRX
 
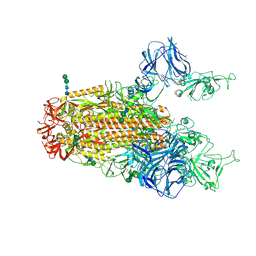 | | SARS Spike Glycoprotein, Stabilized variant, two S1 CTDs in the upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CRW
 
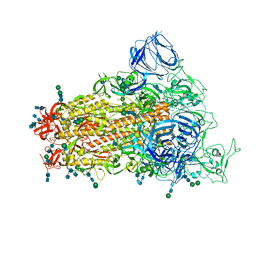 | | SARS Spike Glycoprotein, Stabilized variant, single upwards S1 CTD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CS1
 
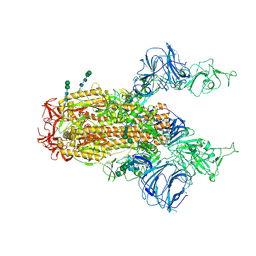 | | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, two S1 CTDs in an upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6OHY
 
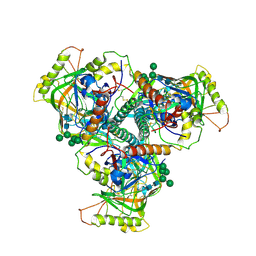 | | Chimpanzee SIV Env trimeric ectodomain. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pallesen, J, Andrabi, R, de Val, N, Burton, D.R, Ward, A.B. | | Deposit date: | 2019-04-08 | | Release date: | 2019-06-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The Chimpanzee SIV Envelope Trimer: Structure and Deployment as an HIV Vaccine Template.
Cell Rep, 27, 2019
|
|
6P62
 
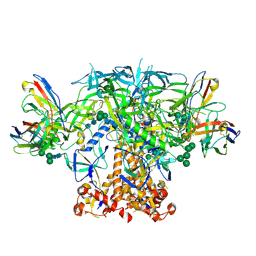 | | HIV Env BG505 NFL TD+ in complex with antibody E70 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env BG505 NFL TD+, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-05-31 | | Release date: | 2019-11-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Vaccination with Glycan-Modified HIV NFL Envelope Trimer-Liposomes Elicits Broadly Neutralizing Antibodies to Multiple Sites of Vulnerability.
Immunity, 51, 2019
|
|
6P8L
 
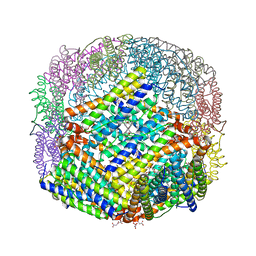 | | Escherichia coli Bacterioferritin Substituted with Zinc Protoporphyrin IX (Zn Absorption Edge X-ray Data) | | Descriptor: | Bacterioferritin, MALONATE ION, PROTOPORPHYRIN IX CONTAINING ZN, ... | | Authors: | Taylor, A.B, Cioloboc, D, Kurtz, D.M. | | Deposit date: | 2019-06-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a Zinc Porphyrin-Substituted Bacterioferritin and Photophysical Properties of Iron Reduction.
Biochemistry, 59, 2020
|
|
6POP
 
 | | Crystal structure of DauA in complex with NADP+ | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase, MAGNESIUM ION, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Insights into the kappa / iota-carrageenan metabolism pathway of some marinePseudoalteromonasspecies.
Commun Biol, 2, 2019
|
|
6OZC
 
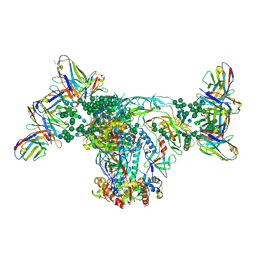 | | BG505 SOSIP.664 with 2G12 Fab2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 Fab Heavy chain, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2019-05-15 | | Release date: | 2020-05-20 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Networks of HIV-1 Envelope Glycans Maintain Antibody Epitopes in the Face of Glycan Additions and Deletions.
Structure, 28, 2020
|
|
