7F7L
 
 | | Crystal structure of AKR4C17 bound with NADPH | | Descriptor: | AKR4-2, COBALT (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7F7K
 
 | | Crystal structure of AKR4C17 bound with NADP+ | | Descriptor: | AKR4-2, COBALT (II) ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7F7J
 
 | | The crystal structure of AKR4C17 | | Descriptor: | AKR4-2, COBALT (II) ION, SULFATE ION | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7T4J
 
 | | Crystal Structure of EGFR_D770_N771insNPG/V948R in complex with TAK-788 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Epidermal growth factor receptor, ... | | Authors: | Skene, R.J, Lane, W, Hu, Y. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of mobocertinib, a potent, oral inhibitor of EGFR exon 20 insertion mutations in non-small cell lung cancer.
Bioorg.Med.Chem.Lett., 80, 2022
|
|
2MYP
 
 | |
2MYT
 
 | | An arsenate reductase in the intermediate state | | Descriptor: | Glutaredoxin arsenate reductase | | Authors: | Jin, C, Yu, C, Hu, C, Hu, Y. | | Deposit date: | 2015-01-30 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A Hybrid Mechanism for the Synechocystis Arsenate Reductase Revealed by Structural Snapshots during Arsenate Reduction.
J.Biol.Chem., 290, 2015
|
|
2MYU
 
 | | An arsenate reductase in oxidized state | | Descriptor: | Glutaredoxin arsenate reductase | | Authors: | Jin, C, Hu, C, Hu, Y. | | Deposit date: | 2015-01-30 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | A Hybrid Mechanism for the Synechocystis Arsenate Reductase Revealed by Structural Snapshots during Arsenate Reduction.
J.Biol.Chem., 290, 2015
|
|
2MYN
 
 | | An arsenate reductase in reduced state | | Descriptor: | Glutaredoxin arsenate reductase | | Authors: | Jin, C, Yu, C, Hu, C, Hu, Y. | | Deposit date: | 2015-01-30 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Hybrid Mechanism for the Synechocystis Arsenate Reductase Revealed by Structural Snapshots during Arsenate Reduction.
J.Biol.Chem., 290, 2015
|
|
1XVA
 
 | | METHYLTRANSFERASE | | Descriptor: | ACETATE ION, GLYCINE N-METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | Authors: | Fu, Z, Hu, Y, Konishi, K, Takata, Y, Ogawa, H, Gomi, T, Fujioka, M, Takusagawa, F. | | Deposit date: | 1996-07-20 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glycine N-methyltransferase from rat liver.
Biochemistry, 35, 1996
|
|
3LKX
 
 | | Human nac dimerization domain | | Descriptor: | Nascent polypeptide-associated complex subunit alpha, Transcription factor BTF3 | | Authors: | Liu, Y, Hu, Y, Li, X, Niu, L, Teng, M. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the human nascent polypeptide-associated complex domain reveals a nucleic acid-binding region on the NACA subunit
Biochemistry, 49, 2010
|
|
4KR0
 
 | | Complex structure of MERS-CoV spike RBD bound to CD26 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Lu, G, Hu, Y, Wang, Q, Qi, J, Gao, F, Li, Y, Zhang, Y, Zhang, W, Yuan, Y, Zhang, B, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-15 | | Release date: | 2013-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26.
Nature, 500, 2013
|
|
4KQZ
 
 | | structure of the receptor binding domain (RBD) of MERS-CoV spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Lu, G, Hu, Y, Wang, Q, Qi, J, Gao, F, Li, Y, Zhang, Y, Zhang, W, Yuan, Y, Bao, J, Zhang, B, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-15 | | Release date: | 2013-07-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.514 Å) | | Cite: | Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26.
Nature, 500, 2013
|
|
1DQI
 
 | | CRYSTAL STRUCTURE OF SUPEROXIDE REDUCTASE FROM P. FURIOSUS IN THE OXIDIZED STATE AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | FE (III) ION, SUPEROXIDE REDUCTASE | | Authors: | Yeh, A.P, Hu, Y, Jenney Jr, F.E, Adams, M.W.W, Rees, D.C. | | Deposit date: | 2000-01-04 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states.
Biochemistry, 39, 2000
|
|
3QWY
 
 | | CED-2 | | Descriptor: | Cell death abnormality protein 2, GLYCEROL, SULFATE ION | | Authors: | Kang, Y, Sun, J, Liu, Y, Sun, D, Hu, Y, Liu, Y.F. | | Deposit date: | 2011-02-28 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of the cell corpse engulfment protein CED-2 in Caenorhabditis elegans.
Biochem.Biophys.Res.Commun., 410, 2011
|
|
1DO6
 
 | | CRYSTAL STRUCTURE OF SUPEROXIDE REDUCTASE IN THE OXIDIZED STATE AT 2.0 ANGSTROM RESOLUTION | | Descriptor: | FE (III) ION, SUPEROXIDE REDUCTASE | | Authors: | Yeh, A.P, Hu, Y, Jenney Junior, F.E, Adams, M.W, Rees, D.C. | | Deposit date: | 1999-12-19 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states.
Biochemistry, 39, 2000
|
|
1DQK
 
 | | CRYSTAL STRUCTURE OF SUPEROXIDE REDUCTASE IN THE REDUCED STATE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | FE (II) ION, SUPEROXIDE REDUCTASE | | Authors: | Yeh, A.P, Hu, Y, Jenney Jr, F.E, Adams, M.W.W, Rees, D.C. | | Deposit date: | 2000-01-04 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states.
Biochemistry, 39, 2000
|
|
4D8M
 
 | | Crystal structure of Bacillus thuringiensis Cry5B nematocidal toxin | | Descriptor: | Pesticidal crystal protein cry5Ba | | Authors: | Fan, H, Hu, Y, Aroian, R.V, Ghosh, P, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2012-01-10 | | Release date: | 2012-12-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Glycolipid Binding Properties of the Nematicidal Protein Cry5B.
Biochemistry, 51, 2012
|
|
8EHK
 
 | |
8EHL
 
 | |
8EHM
 
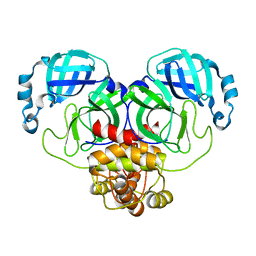 | |
8EHJ
 
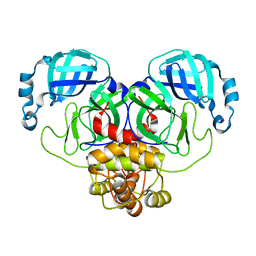 | |
3MOJ
 
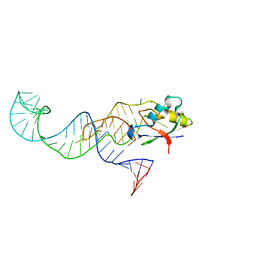 | |
2P0Q
 
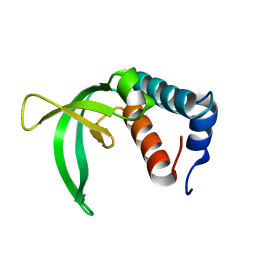 | |
6WOU
 
 | | Cryo-EM structure of recombinant mouse Ryanodine Receptor type 2 mutant R176Q in complex with FKBP12.6 in nanodisc | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Iyer, K.A, Hu, Y, Kurebayashi, N, Murayama, T, Samso, M. | | Deposit date: | 2020-04-25 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural mechanism of two gain-of-function cardiac and skeletal RyR mutations at an equivalent site by cryo-EM.
Sci Adv, 6, 2020
|
|
6WOV
 
 | | Cryo-EM structure of recombinant mouse Ryanodine Receptor type 2 wild type in complex with FKBP12.6 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Iyer, K.A, Hu, Y, Nayak, A.R, Kurebayashi, N, Murayama, T, Samso, M. | | Deposit date: | 2020-04-25 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural mechanism of two gain-of-function cardiac and skeletal RyR mutations at an equivalent site by cryo-EM.
Sci Adv, 6, 2020
|
|
