3M4T
 
 | |
3M1J
 
 | |
3M1R
 
 | | The crystal structure of formimidoylglutamase from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | CACODYLATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tan, K, Bigelow, L, Trevino, D, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-05 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | The crystal structure of formimidoylglutamase from Bacillus subtilis subsp. subtilis str. 168
To be Published
|
|
5AFV
 
 | | Pharmacophore-based virtual screening to discover new active compounds for human choline kinase alpha1. | | Descriptor: | 1,2-ETHANEDIOL, 1-benzyl-4-(benzylamino)-1H-1,2,4-triazol-4-ium, CHOLINE KINASE ALPHA | | Authors: | Serran-Aguilera, L, Nuti, R, Lopez-Cara, L.C, Gallo Mezo, M.A, Macchiarulo, A, Entrena, A, Hurtado-Guerrero, R. | | Deposit date: | 2015-01-26 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Pharmacophore-Based Virtual Screening to Discover New Active Compounds for Human Choline Kinase Alpha1
Mol Inform, 34, 2015
|
|
5AG8
 
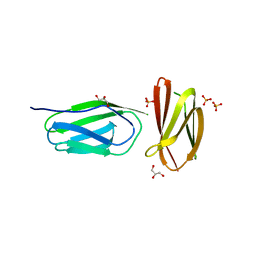 | | CRYSTAL STRUCTURE OF A MUTANT (665I6H) OF THE C-TERMINAL DOMAIN OF RGPB | | Descriptor: | GINGIPAIN R2, GLYCEROL, SULFATE ION | | Authors: | de Diego, I, Ksiazek, M, Mizgalska, D, Golik, P, Szmigielski, B, Nowak, M, Nowakowska, Z, Potempa, B, Koneru, L, Nguyen, K.A, Enghild, J, Thogersen, I.B, Dubin, G, Gomis-Ruth, F.X, Potempa, J. | | Deposit date: | 2015-01-29 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Outer-Membrane Export Signal of Porphyromonas Gingivalis Type Ix Secretion System (T9Ss) is a Conserved C-Terminal Beta-Sandwich Domain.
Sci.Rep., 6, 2016
|
|
4ZXW
 
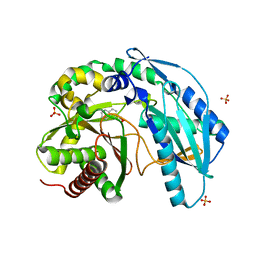 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (complex with (R)-(-)-1-(2-naphthyl)-1,2-ethanediol and sucrose) | | Descriptor: | (1R)-1-(naphthalen-2-yl)ethane-1,2-diol, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, C-domain type II peptide synthetase, ... | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Crystal structure of SgcC5 protein from Streptomyces globisporus
To Be Published
|
|
4ZZF
 
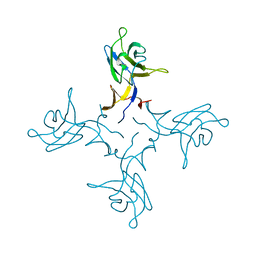 | | Crystal structure of truncated FlgD (tetragonal form) from the human pathogen Helicobacter pylori | | Descriptor: | Flagellar basal body rod modification protein | | Authors: | Pulic, I, Cendron, L, Salamina, M, Matkovic-Calogovic, D, Zanotti, G. | | Deposit date: | 2015-05-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1673 Å) | | Cite: | Crystal structure of truncated FlgD from the human pathogen Helicobacter pylori.
J.Struct.Biol., 194, 2016
|
|
3MDB
 
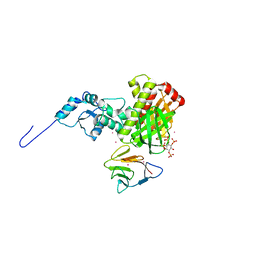 | | Crystal structure of the ternary complex of full length centaurin alpha-1, KIF13B FHA domain, and IP4 | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, Arf-GAP with dual PH domain-containing protein 1, Kinesin-like protein KIF13B, ... | | Authors: | Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-30 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Crystal structure of the ternary complex of full length centaurin alpha-1, KIF13B FHA domain, and IP4
to be published
|
|
4ZW0
 
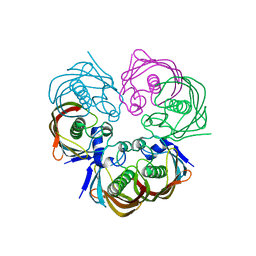 | |
3MEK
 
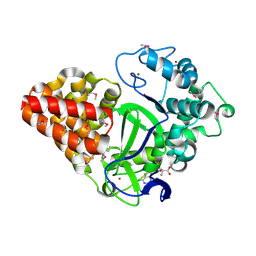 | | Crystal Structure of Human Histone-Lysine N-methyltransferase SMYD3 in Complex with S-adenosyl-L-methionine | | Descriptor: | S-ADENOSYLMETHIONINE, SET and MYND domain-containing protein 3, ZINC ION | | Authors: | Lam, R, Dombrovski, L, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Histone-Lysine N-methyltransferase SMYD3 in Complex with S-adenosyl-L-methionine
To be Published
|
|
3MFM
 
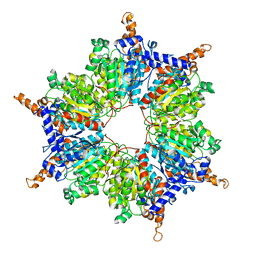 | | Crystal Structures and Mutational Analyses of Acyl-CoA Carboxylase Subunit of Streptomyces coelicolor | | Descriptor: | Propionyl-CoA carboxylase complex B subunit | | Authors: | Diacovich, L, Arabolaza, A, Shillito, E.M, Lin, T.-W, Mitchell, D.L, Melgar, M.M. | | Deposit date: | 2010-04-02 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor.
Biochemistry, 49, 2010
|
|
3MIL
 
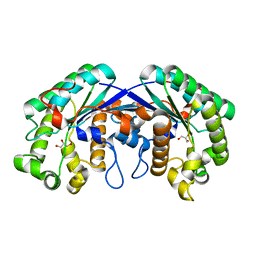 | | Crystal structure of isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Isoamyl acetate-hydrolyzing esterase | | Authors: | Ma, J, Lu, Q, Yuan, Y, Li, K, Ge, H, Go, Y, Niu, L, Teng, M. | | Deposit date: | 2010-04-11 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae reveals a novel active site architecture and the basis of substrate specificity
Proteins, 79, 2011
|
|
5A6U
 
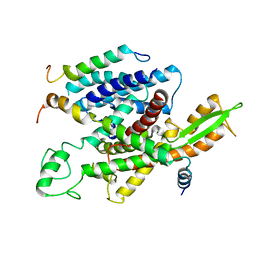 | | Native mammalian ribosome-bound Sec61 protein-conducting channel in the 'non-inserting' state | | Descriptor: | SEC61A, SEC61B, SEC61G | | Authors: | Pfeffer, S, Burbaum, L, Unverdorben, P, Pech, M, Chen, Y, Zimmermann, R, Beckmann, R, Foerster, F. | | Deposit date: | 2015-07-01 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of the Native Sec61 Protein-Conducting Channel.
Nat.Commun., 6, 2015
|
|
5A7U
 
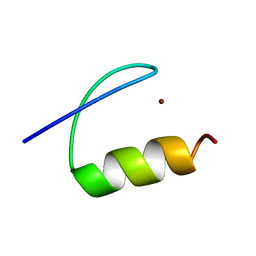 | | Single-particle cryo-EM of co-translational folded adr1 domain inside the E. coli ribosome exit tunnel. | | Descriptor: | REGULATORY PROTEIN ADR1, ZINC ION | | Authors: | Nilsson, O.B, Hedman, R, Marino, J, Wickles, S, Bischoff, L, Johansson, M, Muller-Lucks, A, Trovato, F, Puglisi, J.D, O'Brien, E, Beckmann, R, von Heijne, G. | | Deposit date: | 2015-07-10 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cotranslational Protein Folding Inside the Ribosome Exit Tunnel.
Cell Rep., 12, 2015
|
|
5AEW
 
 | | Crystal structure of II9 variant of Biphenyl dioxygenase from Burkholderia xenovorans LB400 in complex with biphenyl | | Descriptor: | BIPHENYL, BIPHENYL DIOXYGENASE SUBUNIT ALPHA, BIPHENYL DIOXYGENASE SUBUNIT BETA, ... | | Authors: | Dhindwal, S, Gomez-Gil, L, Sylvestre, M, Eltis, L.D, Bolin, J.T, Kumar, P. | | Deposit date: | 2015-01-10 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Basis of the Enhanced Pollutant-Degrading Capabilities of an Engineered Biphenyl Dioxygenase.
J.Bacteriol., 198, 2016
|
|
3MPR
 
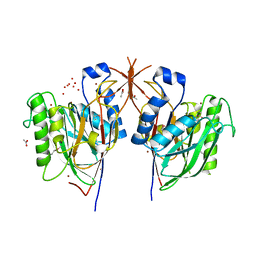 | | Crystal Structure of endonuclease/exonuclease/phosphatase family protein from Bacteroides thetaiotaomicron, Northeast Structural Genomics Consortium Target BtR318A | | Descriptor: | ACETIC ACID, BROMIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-27 | | Release date: | 2010-05-26 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Northeast Structural Genomics Consortium Target BtR318A
To be Published
|
|
5A5U
 
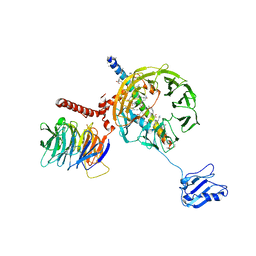 | | Structure of mammalian eIF3 in the context of the 43S preinitiation complex | | Descriptor: | EUKARYOTIC INITIATION FACTOR 3, EUKARYOTIC TRANSLATION INITIATION FACTOR 3 SUBUNIT B, EUKARYOTIC TRANSLATION INITIATION FACTOR 3 SUBUNIT I | | Authors: | des-Georges, A, Dhote, V, Kuhn, L, Hellen, C.U.T, Pestova, T.V, Frank, J, Hashem, Y. | | Deposit date: | 2015-06-21 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of Mammalian Eif3 in the Context of the 43S Preinitiation Complex.
Nature, 525, 2015
|
|
3M1S
 
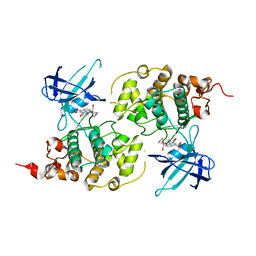 | | Structure of Ruthenium Half-Sandwich Complex Bound to Glycogen Synthase Kinase 3 | | Descriptor: | Glycogen synthase kinase-3 beta, Ruthenium pyridocarbazole | | Authors: | Atilla-Gokcumen, G.E, Di Costanzo, L, Zimmermann, G, Meggers, E. | | Deposit date: | 2010-03-05 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.134 Å) | | Cite: | Structure of anticancer ruthenium half-sandwich complex bound to glycogen synthase kinase 3beta
J.Biol.Inorg.Chem., 16, 2011
|
|
3M3C
 
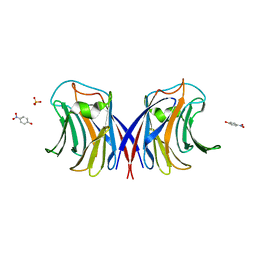 | | Crystal Structure of Agrocybe aegerita lectin AAL complexed with p-Nitrophenyl TF disaccharide | | Descriptor: | Anti-tumor lectin, P-NITROPHENOL, SULFATE ION, ... | | Authors: | Feng, L, Li, D, Wang, D. | | Deposit date: | 2010-03-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the recognition mechanism between an antitumor galectin AAL and the Thomsen-Friedenreich antigen
Faseb J., 24, 2010
|
|
5AKY
 
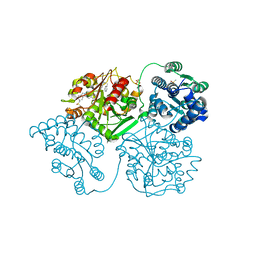 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 3-(2-phenylethyl)-1H-indazole, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-05 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
5A4U
 
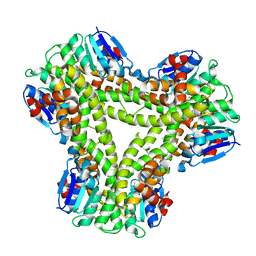 | | AtGSTF2 from Arabidopsis thaliana in complex with indole-3-aldehyde | | Descriptor: | 1H-INDOLE-3-CARBALDEHYDE, ACETATE ION, GLUTATHIONE S-TRANSFERASE F2 | | Authors: | Ahmad, L, Rylott, E, Bruce, N.C, Edwards, R, Grogan, G. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands.
FEBS Open Bio, 7, 2017
|
|
5AKU
 
 | | Crystal structure of TNKS2 in complex with 2-(4-tert-butylphenyl)-1,2, 3,4-tetrahydroquinazolin-4-one | | Descriptor: | 2-(4-tert-butylphenyl)-1,4-dihydroquinazolin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Nkizinkiko, Y, Lehtio, L. | | Deposit date: | 2015-03-05 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Potent and Selective Nonplanar Tankyrase Inhibiting Nicotinamide Mimics.
Bioorg.Med.Chem., 23, 2015
|
|
3M52
 
 | | Crystal structure of the BTB domain from the Miz-1/ZBTB17 transcription regulator | | Descriptor: | ACETATE ION, ZINC ION, Zinc finger and BTB domain-containing protein 17 | | Authors: | Stogios, P.J, Cuesta-Seijo, J.A, Chen, L, Prive, G.G. | | Deposit date: | 2010-03-12 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Insights into Strand Exchange in BTB Domain Dimers from the Crystal Structures of FAZF and Miz1.
J.Mol.Biol., 400, 2010
|
|
5AK5
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 3-PROPYL-5-(3-PYRIDYL)-1H-PYRAZIN-2-ONE, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-02 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
5AK6
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 2-[(5-CHLORO-2-PYRIDYL)SULFANYL]ETHANOL, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-02 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
