3ATO
 
 | | Glycine ethyl ester shielding on the aromatic surfaces of lysozyme: Implication for suppression of protein aggregation | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Ito, L, Shiraki, K, Hasegawa, K, Kumasaka, T. | | Deposit date: | 2011-01-06 | | Release date: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Glycine ethyl ester shielding on the aromatic surfaces of lysozyme: Implication for suppression of protein aggregation
To be Published
|
|
2RN9
 
 | | Solution structure of human apoCox17 | | Descriptor: | Cytochrome c oxidase copper chaperone | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Janicka, A, Martinelli, M, Kozlowski, H, Palumaa, P. | | Deposit date: | 2007-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A structural-dynamical characterization of human cox17
J.Biol.Chem., 283, 2008
|
|
2RNB
 
 | | Solution structure of human Cu(I)Cox17 | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase copper chaperone | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Janicka, A, Martinelli, M, Kozlowski, H, Palumaa, P. | | Deposit date: | 2007-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A structural-dynamical characterization of human cox17
J.Biol.Chem., 283, 2008
|
|
2RLY
 
 | | FBP28WW2 domain in complex with PTPPPLPP peptide | | Descriptor: | Formin-1, Transcription elongation regulator 1 | | Authors: | Ramirez-Espain, X, Ruiz, L, Martin-Malpartida, P, Oschkinat, H, Macias, M.J. | | Deposit date: | 2007-09-03 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a New Binding Motif and a Novel Binding Mode in Group 2 WW Domains
J.Mol.Biol., 373, 2007
|
|
6ZHY
 
 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: hexasome class. | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (110-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Deindl, S. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
3G9H
 
 | | Crystal structure of the C-terminal mu homology domain of Syp1 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Suppressor of yeast profilin deletion | | Authors: | Reider, A, Barker, S, Mishra, S, Im, Y.J, Maldonado-Baez, L, Hurley, J, Traub, L, Wendland, B. | | Deposit date: | 2009-02-13 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation.
Embo J., 28, 2009
|
|
2UUE
 
 | | REPLACE: A strategy for Iterative Design of Cyclin Binding Groove Inhibitors | | Descriptor: | 1-(3,5-DICHLOROPHENYL)-5-METHYL-1H-1,2,4-TRIAZOLE-3-CARBOXYLIC ACID, 4-METHYL-5-{(2E)-2-[(4-MORPHOLIN-4-YLPHENYL)IMINO]-2,5-DIHYDROPYRIMIDIN-4-YL}-1,3-THIAZOL-2-AMINE, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Andrews, M.J, Kontopidis, G, McInnes, C, Plater, A, Innes, L, Cowan, A, Jewsbury, P, Fischer, P.M. | | Deposit date: | 2007-03-02 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Replace: A Strategy for Iterative Design of Cyclin- Binding Groove Inhibitors
Chembiochem, 7, 2006
|
|
2UUR
 
 | | N-terminal NC4 domain of collagen IX | | Descriptor: | 1,2-ETHANEDIOL, COLLAGEN ALPHA-1(IX) CHAIN, SULFATE ION, ... | | Authors: | Leppanen, V.-M, Tossavainen, H, Permi, P, Lehtio, L, Ronnholm, G, Goldman, A, Kilpelainen, I, Pihlajamaa, T. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the N-Terminal Nc4 Domain of Collagen Ix, a Zinc Binding Member of the Laminin-Neurexin-Sex Hormone Binding Globulin (Lns) Domain Family.
J.Biol.Chem., 282, 2007
|
|
2UV6
 
 | | Crystal Structure of a CBS domain pair from the regulatory gamma1 subunit of human AMPK in complex with AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ADENOSINE MONOPHOSPHATE | | Authors: | Day, P, Sharff, A, Parra, L, Cleasby, A, Williams, M, Horer, S, Nar, H, Redemann, N, Tickle, I, Yon, J. | | Deposit date: | 2007-03-09 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Cbs-Domain Pair from the Regulatory Gamma1 Subunit of Human Ampk in Complex with AMP and Zmp.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2UW2
 
 | | Crystal structure of human ribonucleotide reductase subunit R2 | | Descriptor: | FE (III) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE M2 SUBUNIT | | Authors: | Welin, M, Ogg, D, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Kotenyova, T, Magnusdottir, A, Moche, M, Nilsson-Ehle, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Stenmark, P, Uppenberg, J, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Nordlund, P. | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Ribonucleotide Reductase Subunit R2
To be Published
|
|
6XX7
 
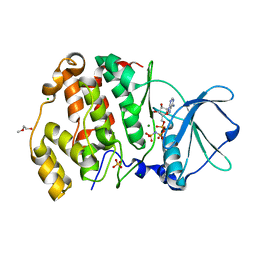 | | Arabidopsis thaliana Casein Kinase 2 (CK2) alpha-1 crystal in complex with ANP | | Descriptor: | CHLORIDE ION, Casein kinase II subunit alpha-1, MAGNESIUM ION, ... | | Authors: | Demulder, M, De Veylder, L, Loris, R. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Arabidopsis thaliana casein kinase 2 alpha 1.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TAV
 
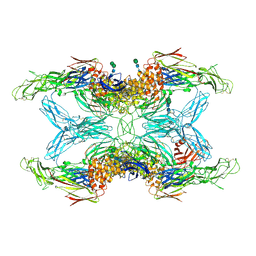 | | Crystal structure of endopeptidase-induced alpha2-macroglobulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin, ... | | Authors: | Gomis-Ruth, F.X, Duquerroy, S, Trapani, S, Marrero, A, Goulas, T, Guevara, T, Andersen, G.A, Sottrup-Jensen, L, Navaza, J. | | Deposit date: | 2019-10-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Cryo-EM structures shows the mechanistic basis of pan-peptidase inhibition by human alpha2-macroglobulin
Proc.Natl.Acad.Sci.USA, 2022
|
|
6Z48
 
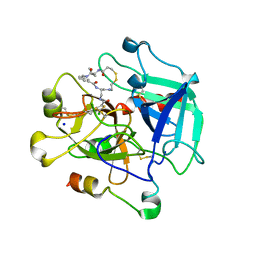 | | Crystal structure of Thrombin in complex with macrocycle X1vE | | Descriptor: | 5-chloranyl-N-[[(4S,15R)-2,5,13,16-tetrakis(oxidanylidene)-15-propan-2-yl-9,10-dithia-3,6,14,17-tetrazabicyclo[17.3.1]tricosa-1(22),19(23),20-trien-4-yl]methyl]thiophene-2-carboxamide, SODIUM ION, Thrombin heavy chain, ... | | Authors: | Angelini, A, Habeshian, S, Heinis, C, Cendron, L. | | Deposit date: | 2020-05-23 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Synthesis and direct assay of large macrocycle diversities by combinatorial late-stage modification at picomole scale.
Nat Commun, 13, 2022
|
|
3BG5
 
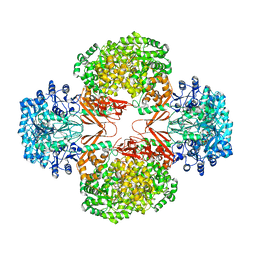 | | Crystal Structure of Staphylococcus Aureus Pyruvate Carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Xiang, S, Tong, L. | | Deposit date: | 2007-11-26 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2UY8
 
 | | R92A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
3BIJ
 
 | | Crystal structure of protein GSU0716 from Geobacter sulfurreducens. Northeast Structural Genomics target GsR13 | | Descriptor: | Uncharacterized protein GSU0716 | | Authors: | Forouhar, F, Neely, H, Su, M, Seetharaman, J, Benach, J, Conover, K, Fang, Y, Xiao, R, Owen, L.A, Maglaqui, M, Cunningham, K, Baran, M.C, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-11 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein GSU0716 from Geobacter sulfurreducens.
To be Published
|
|
2XBV
 
 | | Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor | | Descriptor: | (3R,4R)-1-(2,2-DIFLUORO-ETHYL)-PYRROLIDINE-3,4-DICARBOXYLIC ACID 3-[(5-CHLORO-PYRIDIN-2-YL)-AMIDE]-4-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYL]-AMIDE}, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Banner, D.W, Benz, J, Schlatter, D, Anselm, L, Haap, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of a Factor Xa Inhibitor (3R,4R)-1-(2,2-Difluoro-Ethyl)-Pyrrolidine-3,4-Dicarboxylic Acid 3-[(5-Chloro-Pyridin-2-Yl)-Amide] 4-{[2-Fluoro-4-(2-Oxo-2H-Pyridin-1-Yl)-Phenyl]-Amide} as a Clinical Candidate.
Bioorg.Med.Chem., 20, 2010
|
|
3BJN
 
 | | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae, targeted domain 79-240 | | Descriptor: | CHLORIDE ION, Transcriptional regulator, putative | | Authors: | Chang, C, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae.
To be Published
|
|
2XHD
 
 | | Crystal structure of N-((2S)-5-(6-fluoro-3-pyridinyl)-2,3-dihydro-1H- inden-2-yl)-2-propanesulfonamide in complex with the ligand binding domain of the human GluA2 receptor | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N-[(2S)-5-(6-FLUORO-3-PYRIDINYL)-2,3-DIHYDRO-1H-INDEN-2-YL]-2-PROPANESULFONAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Andreotti, D, Ballantine, S, Bax, B.D, Harris, A.J, Harker, A.J, Lund, J, Melarange, R, Mingardi, A, Mookherjee, C, Mosley, J, Neve, M, Oliosi, B, Profeta, R, Smith, K.J, Smith, P.W, Spada, S, Thewlis, K.M, Yusaf, S.P. | | Deposit date: | 2010-06-14 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of N-[(2S)-5-(6-Fluoro-3-Pyridinyl)-2,3-Dihydro-1H-Inden-2-Yl]-2-Propanesulfonamide, a Novel Clinical Ampa Receptor Positive Modulator.
J.Med.Chem., 53, 2010
|
|
3B2X
 
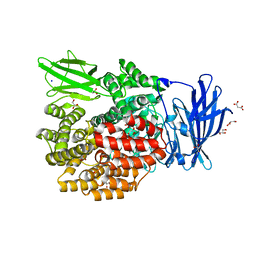 | | Crystal Structure of E. coli Aminopeptidase N in complex with Lysine | | Descriptor: | Aminopeptidase N, GLYCEROL, LYSINE, ... | | Authors: | Addlagatta, A, Gay, L, Matthews, B.W. | | Deposit date: | 2007-10-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the unusual specificity of Escherichia coli aminopeptidase N.
Biochemistry, 47, 2008
|
|
6ZQW
 
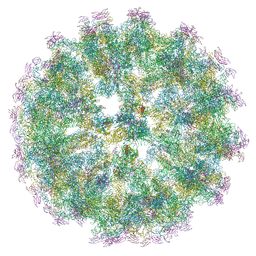 | | Cryo-EM structure of immature Spondweni virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, prM | | Authors: | Renner, M, Dejnirattisai, W, Carrique, L, Serna Martin, I, Karia, D, Ilca, S.L, Ho, S.F, Kotecha, A, Keown, J.R, Mongkolsapaya, J, Screaton, G.R, Grimes, J.M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Nat Commun, 12, 2021
|
|
2XP6
 
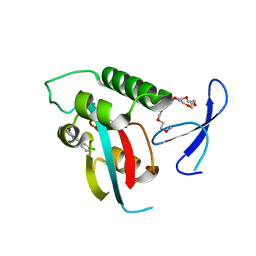 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 2-(3-CHLORO-PHENYL)-5-METHYL-1H-IMIDAZOLE-4-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XFI
 
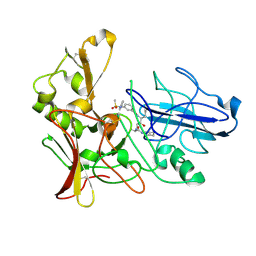 | | Human BACE-1 in complex with N-((1S,2R)-3-(((1S)-2-(cyclohexylamino)- 1-methyl-2-oxoethyl)amino)-2-hydroxy-1-(phenylmethyl)propyl)-3-((methylsulfonyl)(phenyl)amino)benzamide | | Descriptor: | BETA-SECRETASE 1, N-((1S,2R)-3-(((1S)-2-(CYCLOHEXYLAMINO)-1-METHYL-2-OXOETHYL)AMINO)-2-HYDROXY-1-( PHENYLMETHYL)PROPYL)-3-((METHYLSULFONYL)(PHENYL)AMINO) BENZAMIDE | | Authors: | Clarke, B, Cutler, L, Demont, E, Dingwall, C, Dunsdon, R, Hawkins, J, Howes, C, Hussain, I, Maile, G, Matico, R, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2010-05-24 | | Release date: | 2010-07-07 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Bace-1 Inhibitors Using Novel Edge-to-Face Interaction with Arg-296
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XIG
 
 | | The structure of the Helicobacter pylori ferric uptake regulator Fur reveals three functional metal binding sites | | Descriptor: | CITRIC ACID, FERRIC UPTAKE REGULATION PROTEIN, ZINC ION | | Authors: | Dian, C, Vitale, S, Leonard, G.A, Fauquant, F, Muller, C, Bahlawane, C, de Reuse, H, Michaud-Soret, I, Terradot, L. | | Deposit date: | 2010-06-29 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of the Helicobacter Pylori Ferric Uptake Regulator Fur Reveals Three Functional Metal Binding Sites.
Mol.Microbiol., 79, 2011
|
|
3B8T
 
 | | Crystal structure of Escherichia coli alaine racemase mutant P219A | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
