3QP1
 
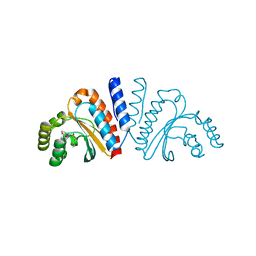 | | Crystal structure of CviR ligand-binding domain bound to the native ligand C6-HSL | | Descriptor: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
1O0F
 
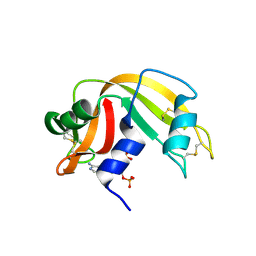 | | RNASE A in complex with 3',5'-ADP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Ribonuclease pancreatic | | Authors: | Leonidas, D.D, Oikonomakos, N.G, Chrysina, E.D, Kosmopoulou, M.N, Vlassi, M. | | Deposit date: | 2003-02-21 | | Release date: | 2003-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structures of ribonuclease A complexed with adenylic and uridylic nucleotide inhibitors. Implications for structure-based design of ribonucleolytic inhibitors
PROTEIN SCI., 12, 2003
|
|
1O0M
 
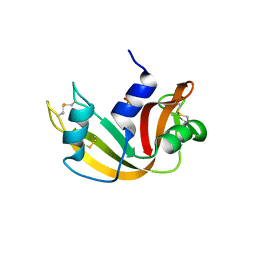 | | Ribonuclease A in complex with uridine-2'-phosphate | | Descriptor: | PHOSPHORIC ACID MONO-[2-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-4-HYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER, Ribonuclease pancreatic | | Authors: | Leonidas, D.D, Oikonomakos, N.G, Chrysina, E.D, Kosmopoulou, M.N, Vlassi, M. | | Deposit date: | 2003-02-24 | | Release date: | 2003-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structures of ribonuclease A complexed with adenylic and uridylic nucleotide inhibitors. Implications for structure-based design of ribonucleolytic inhibitors
PROTEIN SCI., 12, 2003
|
|
1O0N
 
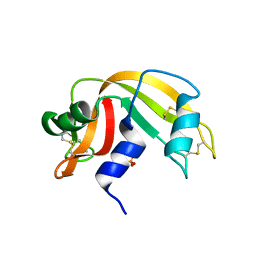 | | Ribonuclease A in complex with uridine-3'-phosphate | | Descriptor: | 3'-URIDINEMONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | Leonidas, D.D, Oikonomakos, N.G, Chrysina, E.D, Kosmopoulou, M.N, Vlassi, M. | | Deposit date: | 2003-02-24 | | Release date: | 2003-12-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structures of ribonuclease A complexed with adenylic and uridylic nucleotide inhibitors. Implications for structure-based design of ribonucleolytic inhibitors
PROTEIN SCI., 12, 2003
|
|
2A0K
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.8 A resolution | | Descriptor: | GLYCEROL, Nucleoside 2-deoxyribosyltransferase, SULFATE ION | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-06-16 | | Release date: | 2005-07-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
1O0O
 
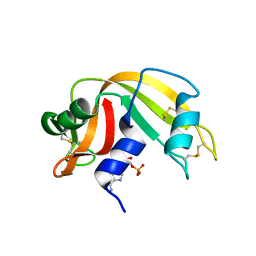 | | Ribonuclease A in complex with adenosine-2',5'-diphosphate | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, Ribonuclease pancreatic | | Authors: | Leonidas, D.D, Oikonomakos, N.G, Chrysina, E.D, Kosmopoulou, M.N, Vlassi, M. | | Deposit date: | 2003-02-24 | | Release date: | 2003-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structures of ribonuclease A complexed with adenylic and uridylic nucleotide inhibitors. Implications for structure-based design of ribonucleolytic inhibitors
PROTEIN SCI., 12, 2003
|
|
2B4T
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum at 2.25 Angstrom resolution reveals intriguing extra electron density in the active site | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Robien, M.A, Bosch, J, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-09-26 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum at 2.25 A resolution reveals intriguing extra electron density in the active site
Proteins, 62, 2006
|
|
1G27
 
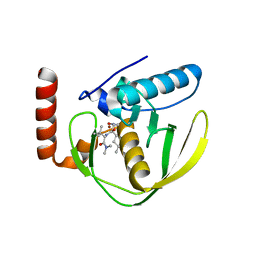 | | CRYSTAL STRUCTURE OF E.COLI POLYPEPTIDE DEFORMYLASE COMPLEXED WITH THE INHIBITOR BB-3497 | | Descriptor: | 2-[(FORMYL-HYDROXY-AMINO)-METHYL]-HEXANOIC ACID (1-DIMETHYLCARBAMOYL-2,2-DIMETHYL-PROPYL)-AMIDE, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-17 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
1G6Y
 
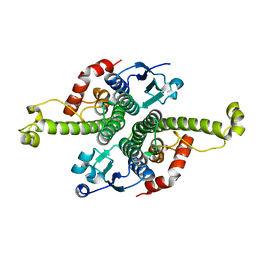 | | CRYSTAL STRUCTURE OF THE GLOBULAR REGION OF THE PRION PROTEIN URE2 FROM YEAST SACCHAROMYCES CEREVISIAE | | Descriptor: | URE2 PROTEIN | | Authors: | Bousset, L, Belrhali, H, Janin, J, Melki, R, Morera, S. | | Deposit date: | 2000-11-08 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the globular region of the prion protein Ure2 from the yeast Saccharomyces cerevisiae.
Structure, 9, 2001
|
|
3FC2
 
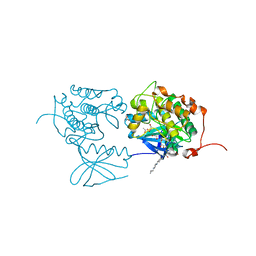 | | PLK1 in complex with BI6727 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bader, G. | | Deposit date: | 2008-11-21 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | BI 6727, A Polo-like Kinase Inhibitor with Improved Pharmacokinetic Profile and Broad Antitumor Activity.
Clin.Cancer Res., 15, 2009
|
|
4BKQ
 
 | | Enoyl-ACP reductase from Yersinia pestis (wildtype)with cofactor NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PUTATIVE REDUCTASE YPZ3_3519 | | Authors: | Hirschbeck, M.W, Neckles, C, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-04-29 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia Pestis Fabv Enoyl-Acp Reductase.
Biochemistry, 55, 2016
|
|
4BKR
 
 | | Enoyl-ACP reductase from Yersinia pestis (wildtype, removed Histag) with cofactor NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hirschbeck, M.W, Neckles, C, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-04-29 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia Pestis Fabv Enoyl-Acp Reductase.
Biochemistry, 55, 2016
|
|
3GI3
 
 | | Crystal structure of a N-Phenyl-N'-Naphthylurea analog in complex with p38 MAP kinase | | Descriptor: | Mitogen-activated protein kinase 14, N-{5-tert-butyl-2-methoxy-3-[({4-[6-(morpholin-4-ylmethyl)pyridin-3-yl]naphthalen-1-yl}carbamoyl)amino]phenyl}methanesulfonamide | | Authors: | Qian, K.C. | | Deposit date: | 2009-03-05 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery and characterization of the N-phenyl-N'-naphthylurea class of p38 kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2HSG
 
 | |
1G6W
 
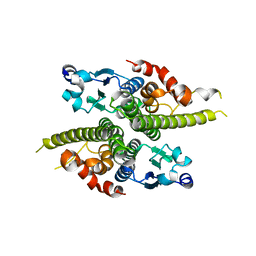 | | CRYSTAL STRUCTURE OF THE GLOBULAR REGION OF THE PRION PROTEIN URE2 FROM THE YEAST SACCAROMYCES CEREVISIAE | | Descriptor: | URE2 PROTEIN | | Authors: | Bousset, L, Belrhali, H, Janin, J, Melki, R, Morera, S. | | Deposit date: | 2000-11-08 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the globular region of the prion protein Ure2 from the yeast Saccharomyces cerevisiae.
Structure, 9, 2001
|
|
1MSC
 
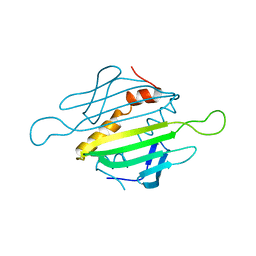 | |
3GYH
 
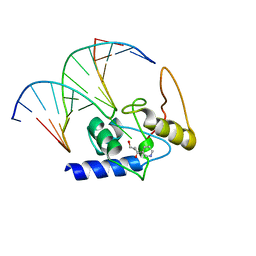 | | Crystal Structure Analysis of S. Pombe ATL in complex with damaged DNA containing POB | | Descriptor: | 1-PYRIDIN-3-YLBUTAN-1-ONE, Alkyltransferase-like protein 1, DNA (5'-D(*CP*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Tubbs, J.L, Arvai, A.S, Tainer, J.A, Shin, D.S. | | Deposit date: | 2009-04-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Flipping of alkylated DNA damage bridges base and nucleotide excision repair.
Nature, 459, 2009
|
|
1G2A
 
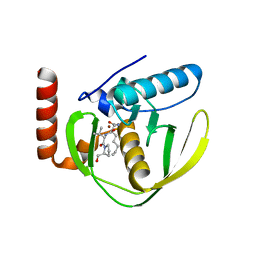 | | THE CRYSTAL STRUCTURE OF E.COLI PEPTIDE DEFORMYLASE COMPLEXED WITH ACTINONIN | | Descriptor: | ACTINONIN, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-18 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
3GVA
 
 | |
3GX4
 
 | | Crystal Structure Analysis of S. Pombe ATL in complex with DNA | | Descriptor: | Alkyltransferase-like protein 1, COBALT HEXAMMINE(III), DNA (5'-D(*CP*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Tubbs, J.L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2009-04-01 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flipping of alkylated DNA damage bridges base and nucleotide excision repair.
Nature, 459, 2009
|
|
4E45
 
 | | Crystal structure of the hMHF1/hMHF2 Histone-Fold Tetramer in Complex with Fanconi Anemia Associated Helicase hFANCM | | Descriptor: | Centromere protein S, Centromere protein X, Fanconi anemia group M protein, ... | | Authors: | Fox III, D, Zhao, Y, Yang, W, Weidong, W. | | Deposit date: | 2012-03-12 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Reveal that FANCM remodels the MHF Tetramer in favor of binding Branched DNA
To be Published
|
|
2Q2A
 
 | | Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus | | Descriptor: | ARGININE, ArtJ, SULFATE ION | | Authors: | Vahedi-Faridi, A, Scheffel, F, Eckey, V, Saenger, W, Schneider, E. | | Deposit date: | 2007-05-26 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP)2
J.Mol.Biol., 375, 2008
|
|
3C4J
 
 | | ABC protein ArtP in complex with ATP-gamma-S | | Descriptor: | Amino acid ABC transporter (ArtP), MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Thaben, P.F, Eckey, V, Scheffel, F, Saenger, W, Schneider, E, Vahedi-Faridi, A. | | Deposit date: | 2008-01-30 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structures of the ATP-binding cassette (ABC) Protein ArtP from Geobacillus stearothermophilus in complexes with nucleotides and nucleotide analogs reveal an intermediate semiclosed dimer in the post hydrolyses state and an asymmetry in the dimerisation region
To be Published
|
|
3CUF
 
 | |
2QYP
 
 | |
