6SM8
 
 | | Human jak1 kinase domain in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-6-[(3~{S})-3-[(1~{S})-2-cyano-1-[4-(7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)pyrazol-1-yl]ethyl]pyrrolidin-1-yl]benzenecarbonitrile, Tyrosine-protein kinase JAK1 | | Authors: | Read, J.A, Steuber, H. | | Deposit date: | 2019-08-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of (2R)-N-[3-[2-[(3-Methoxy-1-methyl-pyrazol-4-yl)amino]pyrimidin-4-yl]-1H-indol-7-yl]-2-(4-methylpiperazin-1-yl)propenamide (AZD4205) as a Potent and Selective Janus Kinase 1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
4GII
 
 | |
7SBQ
 
 | | One RBD-up 1 of pre-fusion SARS-CoV-2 Kappa variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBP
 
 | | Closed state of pre-fusion SARS-CoV-2 Kappa variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBR
 
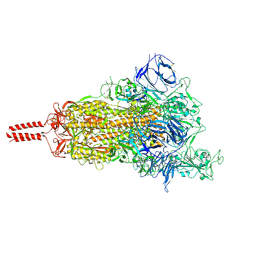 | | One RBD-up 2 of pre-fusion SARS-CoV-2 Kappa variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBO
 
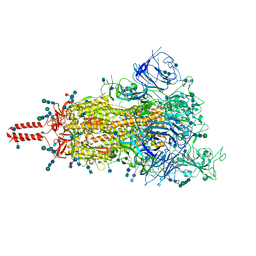 | | One RBD-up 2 of pre-fusion SARS-CoV-2 Delta variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBK
 
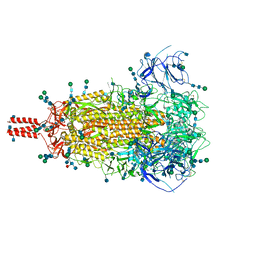 | | Closed state of pre-fusion SARS-CoV-2 Delta variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBS
 
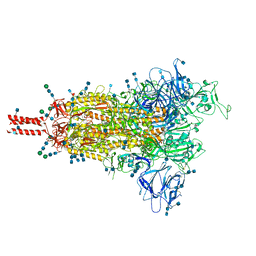 | | One RBD-up 1 of pre-fusion SARS-CoV-2 Gamma variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBT
 
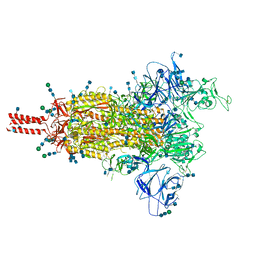 | | One RBD-up 2 of pre-fusion SARS-CoV-2 Gamma variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBL
 
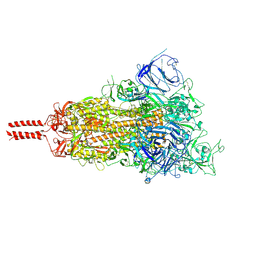 | | One RBD-up 1 of pre-fusion SARS-CoV-2 Delta variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-10 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
4GJ3
 
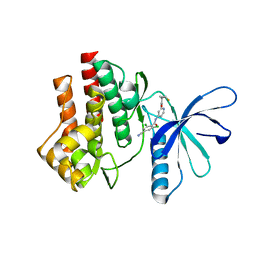 | | Tyk2 (JH1) in complex with 2,6-dichloro-4-cyano-N-[2-({[(1R,2R)-2-fluorocyclopropyl]carbonyl}amino)pyridin-4-yl]benzamide | | Descriptor: | 2,6-dichloro-4-cyano-N-[2-({[(1R,2R)-2-fluorocyclopropyl]carbonyl}amino)pyridin-4-yl]benzamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Ultsch, M.H. | | Deposit date: | 2012-08-09 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lead Optimization of a 4-Aminopyridine Benzamide Scaffold To Identify Potent, Selective, and Orally Bioavailable TYK2 Inhibitors.
J.Med.Chem., 56, 2013
|
|
1GGE
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, NATIVE STRUCTURE AT 1.9 A RESOLUTION. | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, PROTEIN (CATALASE HPII) | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-16 | | Release date: | 2000-08-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
1GGJ
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, ASN201ALA VARIANT. | | Descriptor: | CATALASE HPII, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-21 | | Release date: | 2000-08-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
4R0T
 
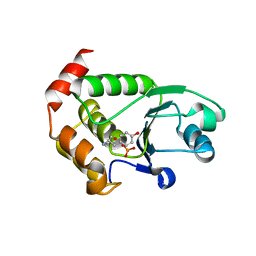 | | Crystal structure of P. aeruginosa TpbA (C132S) in complex with pTyr | | Descriptor: | PHOSPHATE ION, Protein tyrosine phosphatase TpbA, TYROSINE | | Authors: | Xu, K, Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural and Biochemical Analysis of Tyrosine Phosphatase Related to Biofilm Formation A (TpbA) from the Opportunistic Pathogen Pseudomonas aeruginosa PAO1
Plos One, 10
|
|
4R1B
 
 | |
4R0S
 
 | | Crystal structure of P. aeruginosa TpbA | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein tyrosine phosphatase TpbA | | Authors: | Xu, K, Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural and Biochemical Analysis of Tyrosine Phosphatase Related to Biofilm Formation A (TpbA) from the Opportunistic Pathogen Pseudomonas aeruginosa PAO1
PLoS ONE, 10, 2015
|
|
8SZZ
 
 | |
4R1C
 
 | |
4R1A
 
 | |
4R19
 
 | |
5GMZ
 
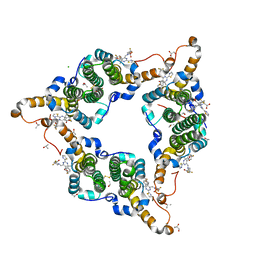 | | Hepatitis B virus core protein Y132A mutant in complex with 4-methyl heteroaryldihydropyrimidine | | Descriptor: | (2S)-4,4-difluoro-1-[[(4S)-4-(4-fluorophenyl)-5-methoxycarbonyl-4-methyl-2-(1,3-thiazol-2-yl)-1H-pyrimidin-6-yl]methyl]pyrrolidine-2-carboxylic acid, CHLORIDE ION, Core protein, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Synthesis of Orally Bioavailable 4-Methyl Heteroaryldihydropyrimidine Based Hepatitis B Virus (HBV) Capsid Inhibitors
J.Med.Chem., 59, 2016
|
|
2YOJ
 
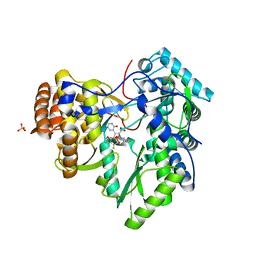 | | HCV NS5B polymerase complexed with pyridonylindole compound | | Descriptor: | 4-fluoranyl-6-[(7-fluoranyl-4-oxidanylidene-3H-quinazolin-6-yl)methyl]-8-(2-oxidanylidene-1H-pyridin-3-yl)furo[2,3-e]indole-7-carboxylic acid, PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Chen, K.X, Venkatraman, S, Anilkumar, G.N, Zeng, Q, Lesburg, C.A, Vibulbhan, B, Yang, W, Velazquez, F, Chan, T.-Y, Bennett, F, Sannigrahi, M, Jiang, Y, Duca, J.S, Pinto, P, Gavalas, S, Huang, Y, Wu, W, Selyutin, O, Agrawal, S, Feld, B, Huang, H.-C, Li, C, Cheng, K.-C, Shih, N.-Y, Kozlowski, J.A, Rosenblum, S.B, Njoroge, F.G. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of Sch 900188: A Potent Hepatitis C Virus Ns5B Polymerase Inhibitor Prodrug as a Development Candidate
Acs Med.Chem.Lett., 5, 2014
|
|
3HI5
 
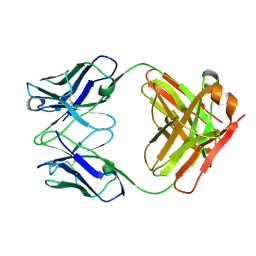 | | Crystal structure of Fab fragment of AL-57 | | Descriptor: | Heavy chain of Fab fragment of AL-57 against alpha L I domain, light chain of Fab fragment of AL-57 against alpha L I domain | | Authors: | Zhang, H, Wang, J. | | Deposit date: | 2009-05-18 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of activation-dependent binding of ligand-mimetic antibody AL-57 to integrin LFA-1.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
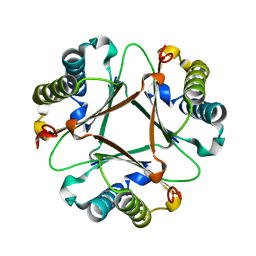
7TVK
 
 | |
8DT0
 
 | |
