7XB1
 
 | | Crystal structure of Omicron BA.3 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, W, Meng, Y, Liao, H. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
8H9Z
 
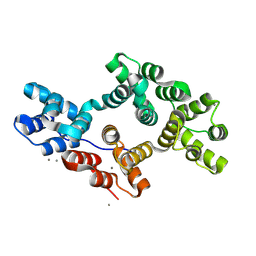 | | Annexin A5 protein mutant | | Descriptor: | Annexin A5, CALCIUM ION | | Authors: | Hua, Z.C, Tang, W. | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-01 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A novel annexin dimer targets microglial phagocytosis of astrocytes to protect the brain-blood barrier after cerebral ischemia.
Acta Pharmacol.Sin., 46, 2025
|
|
5NE4
 
 | |
5NED
 
 | |
7CMZ
 
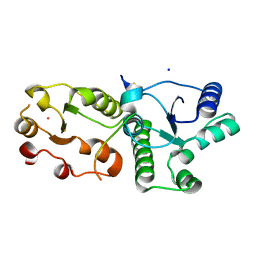 | | Crystal Structure of BRCT7/8 in Complex with the APS Motif of PHF8 | | Descriptor: | DNA topoisomerase 2-binding protein 1, Histone lysine demethylase PHF8, POTASSIUM ION, ... | | Authors: | Che, S.Y, Ma, S, Cao, C, Yao, Z, Shi, L, Yang, N. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | PHF8-promoted TOPBP1 demethylation drives ATR activation and preserves genome stability.
Sci Adv, 7, 2021
|
|
5NEJ
 
 | |
5NET
 
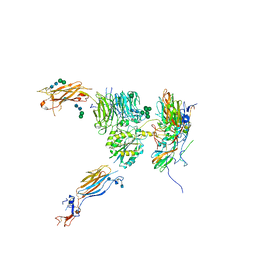 | |
8IFF
 
 | | Cryo-EM structure of Arabidopsis phytochrome A. | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Ma, L, Zhou, C, Wang, J, Guan, Z, Yin, P. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Plant phytochrome A in the Pr state assembles as an asymmetric dimer.
Cell Res., 33, 2023
|
|
5NEU
 
 | |
5NEM
 
 | |
5CNO
 
 | | Crystal structure of the EGFR kinase domain mutant V924R | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Kovacs, E, Das, R, Mirza, A, Jura, N, Barros, T, Kuriyan, J. | | Deposit date: | 2015-07-17 | | Release date: | 2015-07-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Analysis of the Role of the C-Terminal Tail in the Regulation of the Epidermal Growth Factor Receptor.
Mol.Cell.Biol., 35, 2015
|
|
5NER
 
 | |
3M45
 
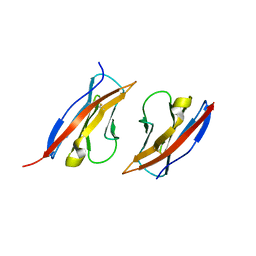 | | Crystal structure of Ig1 domain of mouse SynCAM 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 2 | | Authors: | Yue, L, Modis, Y. | | Deposit date: | 2010-03-10 | | Release date: | 2010-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | N-glycosylation at the SynCAM (synaptic cell adhesion molecule) immunoglobulin interface modulates synaptic adhesion.
J.Biol.Chem., 285, 2010
|
|
5CNN
 
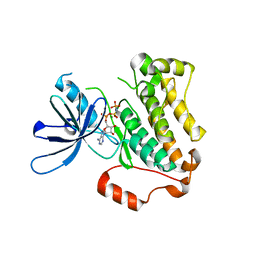 | | Crystal structure of the EGFR kinase domain mutant I682Q | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Kovacs, E, Das, R, Mirza, A, Jura, N, Barros, T, Kuriyan, J. | | Deposit date: | 2015-07-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of the Role of the C-Terminal Tail in the Regulation of the Epidermal Growth Factor Receptor.
Mol.Cell.Biol., 35, 2015
|
|
2PXJ
 
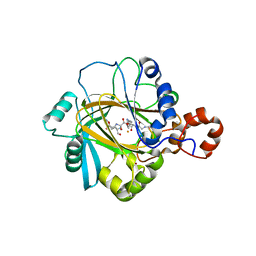 | | The complex structure of JMJD2A and monomethylated H3K36 peptide | | Descriptor: | FE (II) ION, JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, ... | | Authors: | Chen, Z, Zang, J, Kappler, J, Hong, X, Crawford, F, Zhang, G. | | Deposit date: | 2007-05-14 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the recognition of a methylated histone tail by JMJD2A
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2P5B
 
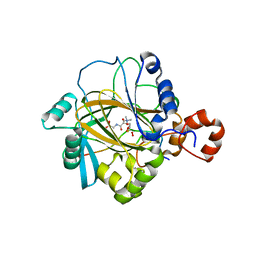 | | The complex structure of JMJD2A and trimethylated H3K36 peptide | | Descriptor: | FE (II) ION, Histone H3, JmjC domain-containing histone demethylation protein 3A, ... | | Authors: | Zhang, G, Chen, Z, Zang, J, Hong, X, Shi, Y. | | Deposit date: | 2007-03-14 | | Release date: | 2007-06-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of the recognition of a methylated histone tail by JMJD2A.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
7WHH
 
 | | Crystal structure of SARS-CoV-2 omicron RBD and human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Wang, X.Q, Lan, J. | | Deposit date: | 2021-12-30 | | Release date: | 2022-06-01 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the SARS-CoV-2 Omicron RBD-ACE2 interaction.
Cell Res., 32, 2022
|
|
8JVA
 
 | | Cryo-EM structure of the N-terminal domain of Omicron BA.1 in complex with nanobody N235 and S2L20 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 heavy chain, S2L20 light chain, ... | | Authors: | Liu, B, Liu, H.H, Han, P, Qi, J.X. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Enhanced potency of an IgM-like nanobody targeting conserved epitope in SARS-CoV-2 spike N-terminal domain.
Signal Transduct Target Ther, 9, 2024
|
|
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24916625 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7EFT
 
 | |
8WU6
 
 | | Structure of a Nerylneryl Diphosphate Synthase from Solanum lycopersicum | | Descriptor: | Nerylneryl diphosphate synthase CPT2, chloroplastic | | Authors: | Li, F.R, Wang, Q.L, Pan, X.M, Dong, L.B. | | Deposit date: | 2023-10-20 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery, Structure, and Engineering of a cis-Geranylfarnesyl Diphosphate Synthase.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8WU7
 
 | | Structure of a cis-Geranylfarnesyl Diphosphate Synthase from Streptomyces clavuligerus | | Descriptor: | Isoprenyl transferase | | Authors: | Li, F.R, Wang, Q.L, Pan, X.M, Dong, L.B. | | Deposit date: | 2023-10-20 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery, Structure, and Engineering of a cis-Geranylfarnesyl Diphosphate Synthase.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YI8
 
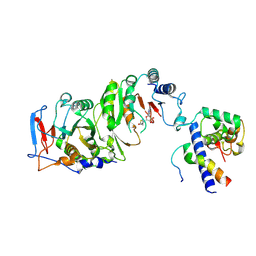 | | Cryo-EM structure of SAH-bound MTA1-MTA9-p1-p2 complex | | Descriptor: | MT-a70 family protein, MTA9, P1, ... | | Authors: | Yan, J.J, Guan, Z.Y, Liu, F.Q, Yan, X.H, Hou, M.J, Yin, P. | | Deposit date: | 2022-07-15 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into DNA N 6 -adenine methylation by the MTA1 complex.
Cell Discov, 9, 2023
|
|
7YI9
 
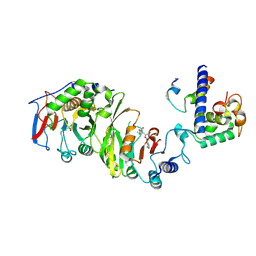 | | Cryo-EM structure of SAM-bound MTA1-MTA9-p1-p2 complex | | Descriptor: | MT-a70 family protein, MTA9, P1, ... | | Authors: | Yan, J.J, Guan, Z.Y, Liu, F.Q, Yan, X.H, Hou, M.J, Yin, P. | | Deposit date: | 2022-07-15 | | Release date: | 2023-01-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into DNA N 6 -adenine methylation by the MTA1 complex.
Cell Discov, 9, 2023
|
|
