3OFN
 
 | | Structure of four mutant forms of yeast F1 ATPase: alpha-N67I | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-15 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
3OU1
 
 | | MDR769 HIV-1 protease complexed with RH/IN hepta-peptide | | Descriptor: | MDR HIV-1 protease, RH/IN substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
7VI7
 
 | | Crystal structure of GH3 beta-N-acetylhexosaminidase Amuc_2109 from Akkermansia muciniphila in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Qian, K, Yang, W, Chen, X, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2021-09-26 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of a GH3 beta-N-acetylhexosaminidase from Akkermansia muciniphila involved in mucin degradation
Biochem.Biophys.Res.Commun., 589, 2022
|
|
7VI6
 
 | | Crystal structure of GH3 beta-N-acetylhexosaminidase Amuc_2109 from Akkermansia muciniphila | | Descriptor: | Beta-N-acetylhexosaminidase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Qian, K, Yang, W, Chen, X, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2021-09-26 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of a GH3 beta-N-acetylhexosaminidase from Akkermansia muciniphila involved in mucin degradation
Biochem.Biophys.Res.Commun., 589, 2022
|
|
8HNS
 
 | | Crystal structure of an anti-CRISPR protein AcrIIC4 in apo form | | Descriptor: | GLYCEROL, anti-CRISPR protein AcrIIC4 | | Authors: | Sun, W, Cheng, Z, Yang, J, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HNW
 
 | | Crystal structure of HpaCas9-sgRNA surveillance complex bound to double-stranded DNA | | Descriptor: | CRISPR-associated endonuclease Cas9, Non-target strand, Target strand, ... | | Authors: | Sun, W, Cheng, Z, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HNT
 
 | |
4KVP
 
 | | Human p53 Core Domain Mutant V157F | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Wallentine, B.D, Wang, Y, Luecke, H. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of oncogenic, suppressor and rescued p53 core-domain variants: mechanisms of mutant p53 rescue.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5Z10
 
 | | Structure of the mechanosensitive Piezo1 channel | | Descriptor: | Piezo-type mechanosensitive ion channel component 1 | | Authors: | Zhao, Q, Zhou, H, Chi, S, Wang, Y, Wang, J, Geng, J, Wu, K, Liu, W, Zhang, T, Dong, M.-Q, Wang, J, Li, X, Xiao, B. | | Deposit date: | 2017-12-22 | | Release date: | 2018-01-31 | | Last modified: | 2020-01-29 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structure and mechanogating mechanism of the Piezo1 channel.
Nature, 554, 2018
|
|
8WQR
 
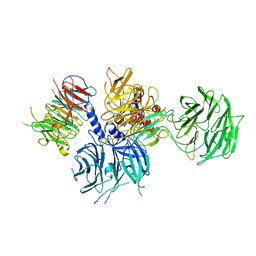 | | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation | | Descriptor: | Activating molecule in BECN1-regulated autophagy protein 1, DNA damage-binding protein 1 | | Authors: | Liu, M, Wang, Y, Su, M.Y, Stjepanovic, G. | | Deposit date: | 2023-10-12 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation.
Nat Commun, 14, 2023
|
|
4KO5
 
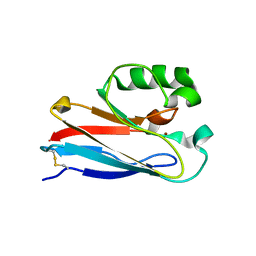 | | Investigating the functional significance of the interlocked pair structural determinants in Pseudomonas aeruginosa azurin (V31I/W48L/V95I/Y108F) | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | Inampudi, K.K, Wang, Y, Meng, W, Tobin, P.H, Wilson, C.J. | | Deposit date: | 2013-05-11 | | Release date: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Investigating the functional significance of the interlocked pair structural determinants in Pseudomonas aeruginosa azurin (V31I/W48L/V95I/Y108F)
To be Published
|
|
7CZ5
 
 | | Cryo-EM structure of the human growth hormone-releasing hormone receptor-Gs protein complex | | Descriptor: | CHOLESTEROL, Growth hormone-releasing hormone receptor,growth hormone-releasing hormone receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhou, F, Zhang, H, Cong, Z, Zhao, L, Zhou, Q, Mao, C, Cheng, X, Shen, D, Cai, X, Ma, C, Wang, Y, Dai, A, Zhou, Y, Sun, W, Zhao, F, Zhao, S, Jiang, H, Jiang, Y, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-07 | | Release date: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for activation of the growth hormone-releasing hormone receptor.
Nat Commun, 11, 2020
|
|
6KZC
 
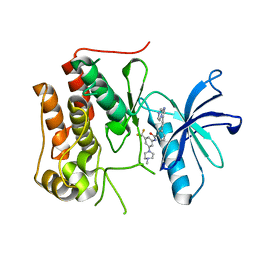 | | crystal structure of TRKc in complex with 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methyl-N-(3-((4- methylpiperazin-1-yl)methyl)-5- (trifluoromethyl)phenyl)benzamide | | Descriptor: | 3-(2-imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methyl-~{N}-[3-(4-methylpiperazin-1-yl)-5-(trifluoromethyl)phenyl]benzamide, NT-3 growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y. | | Deposit date: | 2019-09-23 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis and biological evaluation of 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methylbenzamides as potent and selective pan-tropomyosin receptor kinase (TRK) inhibitors.
Eur.J.Med.Chem., 179, 2019
|
|
7LMA
 
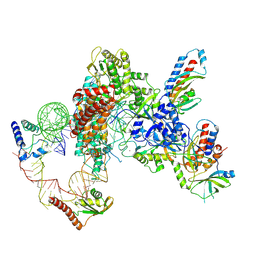 | | Tetrahymena telomerase T3D2 structure at 3.3 Angstrom | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | He, Y, Wang, Y, Liu, B, Helmling, C, Susac, L, Cheng, R, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of telomerase at several steps of telomere repeat synthesis.
Nature, 593, 2021
|
|
7LMB
 
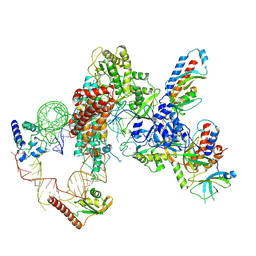 | | Tetrahymena telomerase T5D5 structure at 3.8 Angstrom | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | He, Y, Wang, Y, Liu, B, Helmling, C, Susac, L, Cheng, R, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of telomerase at several steps of telomere repeat synthesis.
Nature, 593, 2021
|
|
4LOF
 
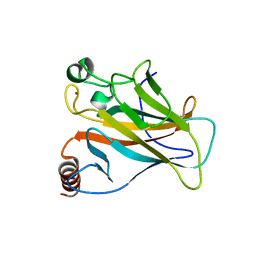 | | Human p53 Core Domain Mutant V157F/N235K/N239Y | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Wallentine, B.D, Wang, Y, Luecke, H. | | Deposit date: | 2013-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of oncogenic, suppressor and rescued p53 core-domain variants: mechanisms of mutant p53 rescue.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4LOE
 
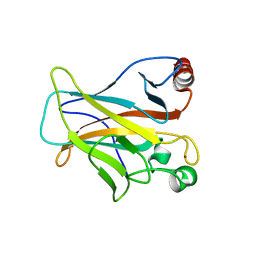 | | Human p53 Core Domain Mutant N239Y | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Wallentine, B.D, Wang, Y, Luecke, H. | | Deposit date: | 2013-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of oncogenic, suppressor and rescued p53 core-domain variants: mechanisms of mutant p53 rescue.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4L1A
 
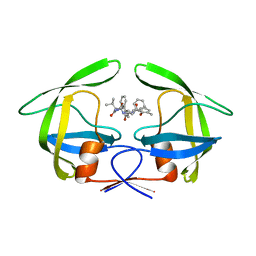 | | Crystallographic study of multi-drug resistant HIV-1 protease Lopinavir complex: mechanism of drug recognition and resistance | | Descriptor: | MDR769 HIV-1 protease, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE | | Authors: | Liu, Z, Yedidi, R.S, Wang, Y, Dewdney, T, Reiter, S, Brunzelle, J, Kovari, I, Kovari, L. | | Deposit date: | 2013-06-03 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study of multi-drug resistant HIV-1 protease lopinavir complex: mechanism of drug recognition and resistance.
Biochem.Biophys.Res.Commun., 437, 2013
|
|
4LO9
 
 | | Human p53 Core Domain Mutant N235K | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Wallentine, B.D, Wang, Y, Luecke, H. | | Deposit date: | 2013-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of oncogenic, suppressor and rescued p53 core-domain variants: mechanisms of mutant p53 rescue.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4EG2
 
 | | 2.2 Angstrom Crystal Structure of Cytidine deaminase from Vibrio cholerae in Complex with Zinc and Uridine | | Descriptor: | ACETATE ION, Cytidine deaminase, MAGNESIUM ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Wang, Y, Grimshaw, S, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Cytidine deaminase from Vibrio cholerae in Complex with Zinc and Uridine.
TO BE PUBLISHED
|
|
8H0O
 
 | | Crystal structure of human serum albumin and ruthenium PZA complex adduct | | Descriptor: | Albumin, CHLORIDE ION, NITRIC OXIDE, ... | | Authors: | Gong, W.J, Wang, Y, Bai, H.H, Wang, W.M, Wang, H.F. | | Deposit date: | 2022-09-30 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.479 Å) | | Cite: | Crystal structure of human serum albumin and ruthenium PZA complex adduct
To Be Published
|
|
4EYR
 
 | | Crystal structure of multidrug-resistant clinical isolate 769 HIV-1 protease in complex with ritonavir | | Descriptor: | HIV-1 PROTEASE, RITONAVIR | | Authors: | Liu, Z, Yedidi, R.S, Wang, Y, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2012-05-01 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of drug resistance: X-ray structure analysis of multi-drug resistant HIV-1 protease ritonavir complex.
Biochem.Biophys.Res.Commun., 431, 2013
|
|
6KH4
 
 | | Design and crystal structure of protein MOFs with ferritin nanocages as linkers and nickel clusters as nodes | | Descriptor: | FE (III) ION, Ferritin, NICKEL (II) ION | | Authors: | Gu, C, Chen, H, Wang, Y, Zhang, T, Wang, H, Zhao, G. | | Deposit date: | 2019-07-12 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural Insight into Binary Protein Metal-Organic Frameworks with Ferritin Nanocages as Linkers and Nickel Clusters as Nodes.
Chemistry, 26, 2020
|
|
4EKU
 
 | |
5XWP
 
 | | Crystal structure of LbuCas13a-crRNA-target RNA ternary complex | | Descriptor: | RNA (30-MER), RNA (59-MER), Uncharacterized protein | | Authors: | Liu, L, Li, X, Li, Z, Wang, Y. | | Deposit date: | 2017-06-30 | | Release date: | 2017-09-13 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a.
Cell, 170, 2017
|
|
