7KFA
 
 | | PCSK9 in complex with PCSK9i a 13mer cyclic peptide LDLR disruptor | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Chopra, R, Xu, M, Spraggon, G. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification of a PCSK9-LDLR disruptor peptide with in vivo function.
Cell Chem Biol, 29, 2022
|
|
7KEV
 
 | | PCSK9 in complex with a cyclic peptide LDLR disruptor | | Descriptor: | CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, Proprotein convertase subtilisin/kexin type 9 Propeptide, ... | | Authors: | Spraggon, G, Chopra, R. | | Deposit date: | 2020-10-12 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a PCSK9-LDLR disruptor peptide with in vivo function.
Cell Chem Biol, 29, 2022
|
|
7ANY
 
 | | Structure of the Laspartomycin C Friulimicin-like mutant in complex with Geranyl phosphate | | Descriptor: | CADMIUM ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zeronian, M.R, Pearce, N.M, Lutz, M, Wood, T.M, Martin, N.I, Janssen, B.J.C. | | Deposit date: | 2020-10-13 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.135 Å) | | Cite: | Mechanistic insights into the C55-P targeting lipopeptide antibiotics revealed by structure-activity studies and high-resolution crystal structures
Chem Sci, 13, 2022
|
|
7AG5
 
 | | Structure of the Laspartomycin C double mutant G4D D-allo-Thr9D-Dap in complex with Geranyl phosphate | | Descriptor: | (~{E})-13-methyltetradec-2-enoic acid, CALCIUM ION, Geranyl phosphate, ... | | Authors: | Zeronian, M.R, Pearce, N.M, Wood, T.M, Martin, N.I, Janssen, B.J.C. | | Deposit date: | 2020-09-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Mechanistic insights into the C55-P targeting lipopeptide antibiotics revealed by structure-activity studies and high-resolution crystal structures
Chem Sci, 13, 2022
|
|
4YW6
 
 | | Structural Insight into Divalent Galactoside Binding to Pseudomonas aeruginosa lectin LecA | | Descriptor: | CALCIUM ION, N-[(2S)-6-amino-1-oxo-1-(pyrrolidin-1-yl)hexan-2-yl]-4-(beta-D-galactopyranosyloxy)benzamide, PA-I galactophilic lectin | | Authors: | Visini, R, Jin, X, Michaud, G, Bergmann, M, Gillon, E, Imberty, A, Stocker, A, Darbre, T, Pieters, R, Reymond, J.-L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Insight into Multivalent Galactoside Binding to Pseudomonas aeruginosa Lectin LecA.
Acs Chem.Biol., 10, 2015
|
|
5D6J
 
 | | Crystal structure of a mycobacterial protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-CoA synthase, MAGNESIUM ION, ... | | Authors: | Li, W.J, Bi, L.J. | | Deposit date: | 2015-08-12 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of FadD32, an enzyme essential for mycolic acid biosynthesis in mycobacteria.
Sci Rep, 5, 2015
|
|
5D6N
 
 | | Crystal structure of a mycobacterial protein | | Descriptor: | Acyl-CoA synthase | | Authors: | Li, W.J, Bi, L.J. | | Deposit date: | 2015-08-12 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of FadD32, an enzyme essential for mycolic acid biosynthesis in mycobacteria.
Sci Rep, 5, 2015
|
|
4YW7
 
 | | Structural Insight into Divalent Galactoside Binding to Pseudomonas aeruginosa lectin LecA | | Descriptor: | (2R,3R,4S,5R,6R,2'R,3'R,4'S,5'R,6'R)-2,2'-([(2R,3R,4S,5S,6S)-3,4-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2,5-diyl]bis{1H-1,2,3-triazole-1,4-diyl[(2S,3R,4S,5S,6S)-3,4-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2,5-diyl]-1H-1,2,3-triazole-1,4-diylpropane-3,1-diyloxy})bis[6-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol], CALCIUM ION, PA-I galactophilic lectin, ... | | Authors: | Visini, R, Jin, X, Michaud, G, Bergmann, M, Gillon, E, Imberty, A, Stocker, A, Darbre, T, Pieters, R, Reymond, J.-L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Insight into Multivalent Galactoside Binding to Pseudomonas aeruginosa Lectin LecA.
Acs Chem.Biol., 10, 2015
|
|
4YWA
 
 | | Structural Insight into Divalent Galactoside Binding to Pseudomonas aeruginosa lectin LecA | | Descriptor: | (2R,3R,4S,5R,6R,2'R,3'R,4'S,5'R,6'R)-2,2'-([(2R,3R,4S,5S,6S)-3,4-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2,5-diyl]bis{1H-1,2,3-triazole-1,4-diyl[(2S,3R,4S,5S,6S)-3,4-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2,5-diyl]-1H-1,2,3-triazole-1,4-diylmethanediyloxy})bis[6-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol], CALCIUM ION, PA-I galactophilic lectin | | Authors: | Visini, R, Jin, X, Michaud, G, Bergmann, M, Gillon, E, Imberty, A, Stocker, A, Darbre, T, Pieters, R, Reymond, J.-L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.192 Å) | | Cite: | Structural Insight into Multivalent Galactoside Binding to Pseudomonas aeruginosa Lectin LecA.
Acs Chem.Biol., 10, 2015
|
|
3L59
 
 | | Structure of BACE Bound to SCH710413 | | Descriptor: | (2Z)-3-(3-chlorobenzyl)-2-imino-5,5-dimethylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L58
 
 | | Structure of BACE Bound to SCH589432 | | Descriptor: | Beta-secretase 1, N'-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-5-METHYL-N,N-DIPROPYLISOPHTHALAMIDE | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5B
 
 | | Structure of BACE Bound to SCH713601 | | Descriptor: | (2Z,5R)-3-(3-chlorobenzyl)-2-imino-5-methyl-5-(2-methylpropyl)imidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5C
 
 | | Structure of BACE Bound to SCH723871 | | Descriptor: | 1-(4-cyanophenyl)-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
8SPB
 
 | | Caspase-4/Pro-IL-18 complex | | Descriptor: | Caspase-4 subunit p10, Caspase-4 subunit p20, Interleukin-18 | | Authors: | Pascal, D, Dong, Y, Wu, H, Jon, K. | | Deposit date: | 2023-05-02 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into cytokine cleavage by inflammatory caspase-4.
Nature, 624, 2023
|
|
3L5F
 
 | | Structure of BACE Bound to SCH736201 | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-5-(cyclohexylmethyl)-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3CIC
 
 | | Structure of BACE Bound to SCH709583 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2S)-2-[(2S)-4-benzyl-3-oxopiperazin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7KLM
 
 | | Human Arginase1 Complexed with Inhibitor Compound 24a | | Descriptor: | 3-[(2~{S},3~{R},4~{R})-4-[[(2~{S})-2-azanyl-3-methyl-butanoyl]amino]-2-carboxy-pyrrolidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boron, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-Based Discovery of Proline-Derived Arginase Inhibitors with Improved Oral Bioavailability for Immuno-Oncology.
Acs Med.Chem.Lett., 12, 2021
|
|
7KLL
 
 | | Human Arginase1 Complexed with Inhibitor Compound 18 | | Descriptor: | 3-[(2~{S},3~{R},4~{R})-4-azanyl-2-carboxy-pyrrolidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boron, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-Based Discovery of Proline-Derived Arginase Inhibitors with Improved Oral Bioavailability for Immuno-Oncology.
Acs Med.Chem.Lett., 12, 2021
|
|
7KLK
 
 | | Human Arginase1 Complexed with Inhibitor Compound 3a | | Descriptor: | 1,2-ETHANEDIOL, 3-[(2~{S},3~{R})-2-carboxypiperidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, ... | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure-Based Discovery of Proline-Derived Arginase Inhibitors with Improved Oral Bioavailability for Immuno-Oncology.
Acs Med.Chem.Lett., 12, 2021
|
|
3CIB
 
 | | Structure of BACE Bound to SCH727596 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2R)-2-[(2R,4S)-4-benzylpiperidin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3L5D
 
 | | Structure of BACE Bound to SCH723873 | | Descriptor: | 1-butyl-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3CID
 
 | | Structure of BACE Bound to SCH726222 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2S)-2-[(4S)-1-benzyl-5-oxoimidazolidin-4-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4DJV
 
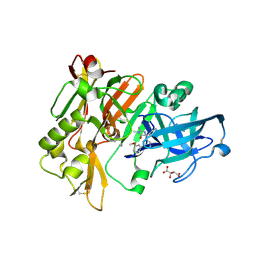 | | Structure of BACE Bound to 2-imino-5-(3'-methoxy-[1,1'-biphenyl]-3-yl)-3-methyl-5-phenylimidazolidin-4-one | | Descriptor: | (2E,5R)-2-imino-5-(3'-methoxybiphenyl-3-yl)-3-methyl-5-phenylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-21 | | Last modified: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure based design of iminohydantoin BACE1 inhibitors: Identification of an orally available, centrally active BACE1 inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6KC0
 
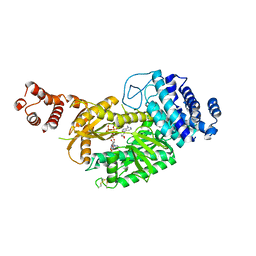 | | fused To-MtbCsm1 with 2ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A),CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), MAGNESIUM ION | | Authors: | Li, T, Huo, Y, Jiang, T. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Mycobacterium tuberculosis CRISPR/Cas system Csm1 holds clues to the evolutionary relationship between DNA polymerase and cyclase activity.
Int.J.Biol.Macromol., 170, 2020
|
|
4DJY
 
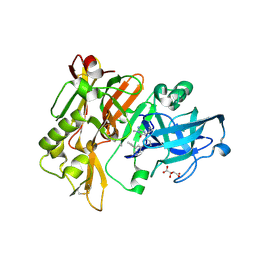 | |
