7YVM
 
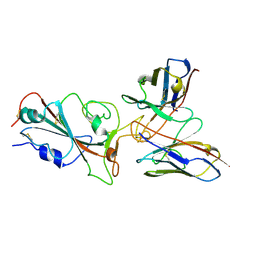 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH272 Fab | | Descriptor: | Spike glycoprotein, TH281 Fab heavy chain, TH281 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVO
 
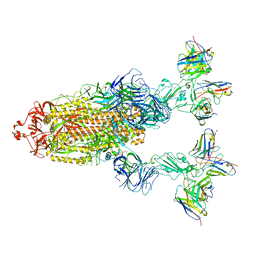 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH027/132 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH132 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVH
 
 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH132 Fab | | Descriptor: | Spike glycoprotein, TH132 Fab heavy chain, TH132 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVF
 
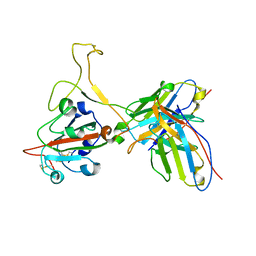 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH027 Fab | | Descriptor: | Spike glycoprotein, TH027 Fab heavy chain, TH027 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVJ
 
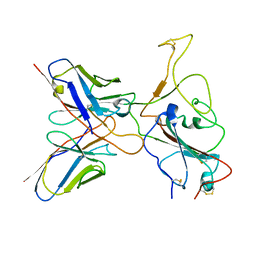 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH236 Fab | | Descriptor: | Spike glycoprotein, TH236 Fab heavy chain, TH236 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVL
 
 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH272 Fab | | Descriptor: | Spike glycoprotein, TH272 Fab heavy chain, TH272 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVI
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH236 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH236 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
6J1Q
 
 | | Crystal structure of Candida Antarctica Lipase B mutant - RS | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
3OBT
 
 | |
3NMX
 
 | | Crystal structure of APC complexed with Asef | | Descriptor: | APC variant protein, Rho guanine nucleotide exchange factor 4 | | Authors: | Zhang, Z, Chen, L, Gao, L, Lin, K, Wu, G. | | Deposit date: | 2010-06-22 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the recognition of Asef by adenomatous polyposis coli.
Cell Res., 22, 2012
|
|
4EQK
 
 | |
4ENF
 
 | | Crystal structure of the cap-binding domain of polymerase basic protein 2 from influenza virus A/Puerto Rico/8/34(h1n1) | | Descriptor: | 1,4-BUTANEDIOL, NITRATE ION, Polymerase basic protein 2 | | Authors: | Meng, G, Liu, Y, Zheng, X. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural and functional characterization of K339T substitution identified in the PB2 subunit cap-binding pocket of influenza A virus
J.Biol.Chem., 288, 2013
|
|
4ISH
 
 | | Structure of FACTOR VIIA in complex with the inhibitor BMS-593214 also known as 2'-[(6R,6AR,11BR)-2-CARBAMIMIDOYL-6,6A,7,11B-TETRAHYDRO-5H-INDENO[2,1-C]QUINOLIN-6-YL]-5'-HYDROXY-4'-METHOXYBIPHENYL-4-CARBOXYLIC ACID | | Descriptor: | 2'-[(6R,6aR,11bR)-2-carbamimidoyl-6,6a,7,11b-tetrahydro-5H-indeno[2,1-c]quinolin-6-yl]-5'-hydroxy-4'-methoxybiphenyl-4-carboxylic acid, CALCIUM ION, Factor VII heavy chain, ... | | Authors: | Wei, A. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery and gram-scale synthesis of BMS-593214, a potent, selective FVIIa inhibitor.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3NMZ
 
 | | Crystal structure of APC complexed with Asef | | Descriptor: | APC variant protein, Rho guanine nucleotide exchange factor 4 | | Authors: | Zhang, Z, Chen, L, Gao, L, Lin, K, Wu, G. | | Deposit date: | 2010-06-23 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for the recognition of Asef by adenomatous polyposis coli.
Cell Res., 22, 2012
|
|
3WY7
 
 | | Crystal structure of Mycobacterium smegmatis 7-Keto-8-aminopelargonic acid (KAPA) synthase BioF | | Descriptor: | 8-amino-7-oxononanoate synthase | | Authors: | Fan, S.H, Li, D.F, Wang, D.C, Chen, G.J, Zhang, X.E, Bi, L.J. | | Deposit date: | 2014-08-20 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of Mycobacterium smegmatis 7-keto-8-aminopelargonic acid (KAPA) synthase
Int.J.Biochem.Cell Biol., 58C, 2014
|
|
4JV7
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JVR
 
 | | Co-crystal structure of MDM2 with inhibitor (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide | | Descriptor: | (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-26 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JV9
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
7DWB
 
 | | Human Pannexin1 model | | Descriptor: | Pannexin-1 | | Authors: | Zhang, S.S, Yang, M.J. | | Deposit date: | 2021-01-17 | | Release date: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structure of the full-length human Pannexin1 channel and insights into its role in pyroptosis.
Cell Discov, 7, 2021
|
|
4JVE
 
 | | Co-crystal structure of MDM2 with inhibitor (2R,3E)-2-[(2S,3R,6S)-2,3-bis(4-chlorophenyl)-6-(4-fluorobenzyl)-5-oxomorpholin-4-yl]pent-3-enoic acid | | Descriptor: | (2R,3E)-2-[(2S,3R,6S)-2,3-bis(4-chlorophenyl)-6-(4-fluorobenzyl)-5-oxomorpholin-4-yl]pent-3-enoic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4ES5
 
 | |
3Q96
 
 | | B-Raf kinase domain in complex with a tetrahydronaphthalene inhibitor | | Descriptor: | (2S)-N-[3-(2-aminopropan-2-yl)-5-(trifluoromethyl)phenyl]-7-[(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-4-yl)oxy]-1,2,3,4-tetrahydronaphthalene-2-carboxamide, Serine/threonine-protein kinase B-raf | | Authors: | Sintchak, M.D, Aertgeerts, K, Yano, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Design and optimization of potent and orally bioavailable tetrahydronaphthalene raf inhibitors.
J.Med.Chem., 54, 2011
|
|
6IFE
 
 | | A Glycoside Hydrolase Family 43 beta-Xylosidase | | Descriptor: | Beta-xylosidase, GLYCEROL | | Authors: | Li, N, Liu, Y, Zhang, R, Zhou, J.P, Huang, Z.X. | | Deposit date: | 2018-09-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Biochemical and structural properties of a low-temperature-active glycoside hydrolase family 43 beta-xylosidase: Activity and instability at high neutral salt concentrations.
Food Chem, 301, 2019
|
|
6SS4
 
 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0021181 | | Descriptor: | Arginase-2, mitochondrial, Fab C0021181 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS2
 
 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0021158 | | Descriptor: | Arginase-2, mitochondrial, Fab C0021158 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
