8XDD
 
 | | Cryo-EM structure of human urea transporter A2. | | Descriptor: | 1-(3-methoxyphenyl)methanamine, 8-hydroxyquinoline-2-carboxylic acid, Urea transporter 2 | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
2J5F
 
 | |
7WUJ
 
 | | Tethered peptide activation mechanism of adhesion GPCRs ADGRG2 and ADGRG4 | | Descriptor: | Adhesion G-protein coupled receptor G4,Uncharacterized protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Guo, S.C, Huang, S.M, He, Q.T, Xiao, P, Sun, J.P, Yu, X. | | Deposit date: | 2022-02-08 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4.
Nature, 604, 2022
|
|
7M8W
 
 | | XFEL crystal structure of the prostaglandin D2 receptor CRTH2 in complex with 15R-methyl-PGD2 | | Descriptor: | 15R-methyl-prostaglandin D2, CITRATE ANION, Prostaglandin D2 receptor 2, ... | | Authors: | Shiriaeva, A, Han, G.W, Cherezov, V. | | Deposit date: | 2021-03-30 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Molecular basis for lipid recognition by the prostaglandin D 2 receptor CRTH2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2J5E
 
 | |
7VKK
 
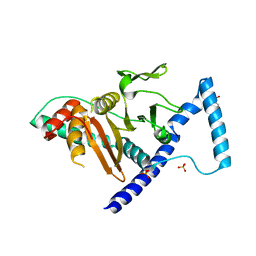 | | Crystal structure of D. melanogaster SAMTOR V66W/E67P mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-adenosylmethionine sensor upstream of mTORC1, SULFATE ION | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Molecular mechanism of S -adenosylmethionine sensing by SAMTOR in mTORC1 signaling.
Sci Adv, 8, 2022
|
|
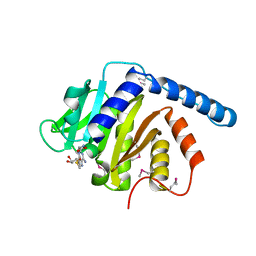
7VKR
 
 | |
7VKQ
 
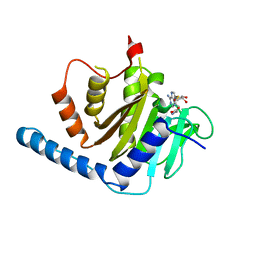 | |
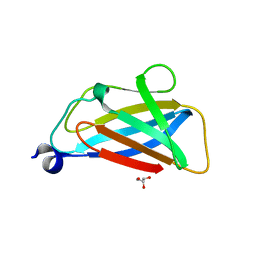
5TUH
 
 | |
7WUI
 
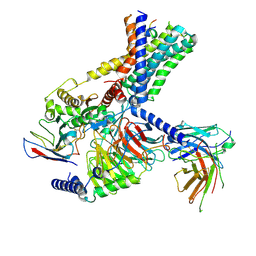 | | Tethered peptide activation mechanism of adhesion GPCRs ADGRG2 and ADGRG4 | | Descriptor: | Adhesion G-protein coupled receptor G2,mCherry, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Guo, S.C, He, Q.T, Xiao, P, Sun, J.P, Yu, X. | | Deposit date: | 2022-02-08 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4.
Nature, 604, 2022
|
|
5TUG
 
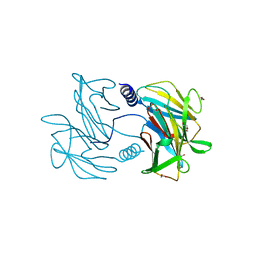 | |
6IPP
 
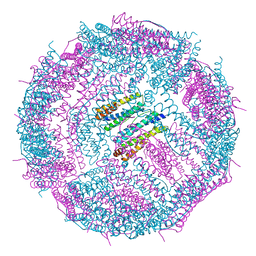 | | Non-native ferritin 8-mer mutant-C90A/C102A/C130A/D144C | | Descriptor: | FE (III) ION, Ferritin heavy chain | | Authors: | Zang, J, Chen, H, Zhou, K, Zhao, G. | | Deposit date: | 2018-11-03 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Disulfide-mediated conversion of 8-mer bowl-like protein architecture into three different nanocages.
Nat Commun, 10, 2019
|
|
6JC7
 
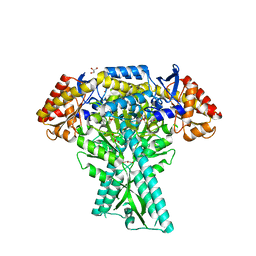 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with amino donor L-Ala | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-alanine, ACETIC ACID, CrmG, ... | | Authors: | Xu, J, Su, K, Liu, J. | | Deposit date: | 2019-01-28 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies reveal flexible roof of active site responsible for omega-transaminase CrmG overcoming by-product inhibition.
Commun Biol, 3, 2020
|
|
8XKN
 
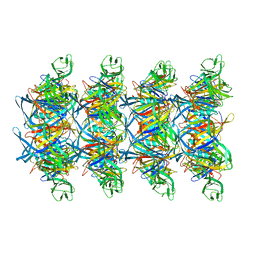 | | Cryo-EM structure of tail tube protein | | Descriptor: | a protein | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-12-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
6JC9
 
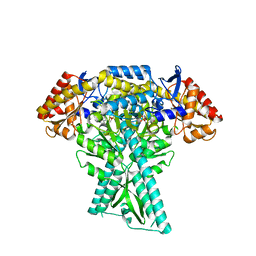 | |
6JLC
 
 | |
7DGB
 
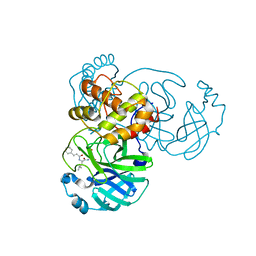 | |
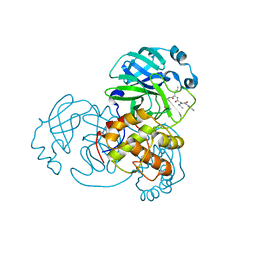
7DGH
 
 | |
7DGF
 
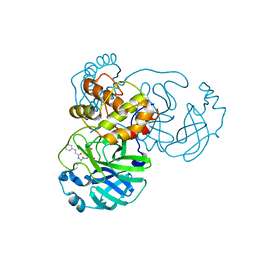 | |
7DGG
 
 | |
7DGI
 
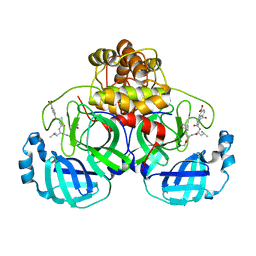 | |
7DHJ
 
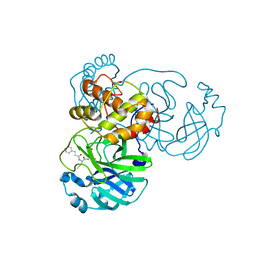 | | The co-crystal structure of SARS-CoV-2 main protease with the peptidomimetic inhibitor (S)-2-cinnamamido-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)pent-4-ynamide | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pent-4-ynamide, 3C-like proteinase | | Authors: | Shang, L.Q, Wang, H, Deng, W.L, Xing, S, Wang, Y.X. | | Deposit date: | 2020-11-15 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | The structure-based design of peptidomimetic inhibitors against SARS-CoV-2 3C like protease as Potent anti-viral drug candidate.
Eur.J.Med.Chem., 238, 2022
|
|
6IPQ
 
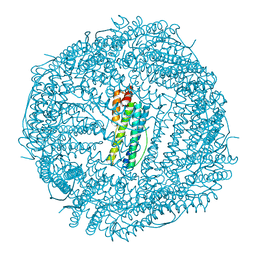 | | Non-native ferritin 8-mer mutant-C90A/C102A/C130A | | Descriptor: | Ferritin heavy chain, MAGNESIUM ION | | Authors: | Zang, J, Chen, H, Wang, Y, Zhao, G. | | Deposit date: | 2018-11-03 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Disulfide-mediated conversion of 8-mer bowl-like protein architecture into three different nanocages.
Nat Commun, 10, 2019
|
|
8EWG
 
 | | Cryo-EM structure of a riboendonclease | | Descriptor: | CRISPR-associated endonuclease Cas9, RNA (56-MER) | | Authors: | Li, Z, Wang, F. | | Deposit date: | 2022-10-23 | | Release date: | 2023-08-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Basis for the Ribonuclease Activity of a Thermostable CRISPR-Cas13a from Thermoclostridium caenicola.
J.Mol.Biol., 435, 2023
|
|
5F9P
 
 | | Crystal structure study of anthrone oxidase-like protein | | Descriptor: | Anthrone oxidase-like protein, GLYCEROL | | Authors: | Gao, X, Wu, D, Fan, K, Liu, Z.-J. | | Deposit date: | 2015-12-10 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.078 Å) | | Cite: | Structure and Function of a C-C Bond Cleaving Oxygenase in Atypical Angucycline Biosynthesis
ACS Chem. Biol., 12, 2017
|
|
