7LAQ
 
 | |
6CF9
 
 | |
6OAP
 
 | | Crystal structure of a dual sensor histidine kinase in the green-light absorbing Pg state | | Descriptor: | Dual sensor histidine kinase, MAGNESIUM ION, PHYCOCYANOBILIN | | Authors: | Heewhan, S, Zhong, R, Xiaoli, Z, Sepalika, B, Xiaojing, Y. | | Deposit date: | 2019-03-18 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of molecular logic OR in a dual-sensor histidine kinase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OAQ
 
 | | Crystal structure of a dual sensor histidine kinase in BeF3- bound state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Dual sensor histidine kinase, MAGNESIUM ION, ... | | Authors: | Heewhan, S, Zhong, R, Xiaoli, Z, Sepalika, B, Xiaojing, Y. | | Deposit date: | 2019-03-18 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural basis of molecular logic OR in a dual-sensor histidine kinase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OB8
 
 | | Crystal structure of a dual sensor histidine kinase in green light illuminated state | | Descriptor: | Dual sensor histidine kinase, MAGNESIUM ION, PHYCOCYANOBILIN | | Authors: | Heewhan, S, Zhong, R, Xiaoli, Z, Sepalika, B, Xiaojing, Y. | | Deposit date: | 2019-03-19 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of molecular logic OR in a dual-sensor histidine kinase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OZB
 
 | | Crystal structure of the phycoerythrobilin-bound GAF domain from a cyanobacterial phytochrome | | Descriptor: | PHYCOERYTHROBILIN, Two-component sensor histidine kinase | | Authors: | Heewhan, S, Xiaoli, Z, Yafang, S, Zhong, R, Wolfgang, G, Kai, H.Z, Xiaojing, Y. | | Deposit date: | 2019-05-15 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The interplay between chromophore and protein determines the extended excited state dynamics in a single-domain phytochrome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OZA
 
 | | Crystal structure of the phycocyanobilin-bound GAF domain from a cyanobacterial phytochrome | | Descriptor: | PHYCOCYANOBILIN, Two-component sensor histidine kinase | | Authors: | Heewhan, S, Xiaoli, Z, Yafang, S, Zhong, R, Wolfgang, G, Kai, H.Z, Xiaojing, Y. | | Deposit date: | 2019-05-15 | | Release date: | 2020-05-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | The interplay between chromophore and protein determines the extended excited state dynamics in a single-domain phytochrome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7RWK
 
 | | Structure of Cap5 from Asticcacaulis sp. | | Descriptor: | MAGNESIUM ION, SAVED domain-containing protein, ZINC ION | | Authors: | Huang, R.H, Fatma, S, Chakravarti, A. | | Deposit date: | 2021-08-20 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Molecular mechanisms of the CdnG-Cap5 antiphage defense system employing 3',2'-cGAMP as the second messenger.
Nat Commun, 12, 2021
|
|
7RWM
 
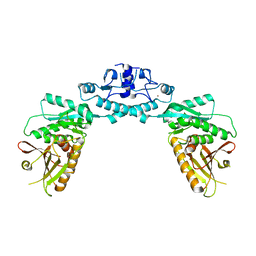 | | Structure of Cap5 from Lactococcus lactis | | Descriptor: | SAVED domain-containing protein, ZINC ION | | Authors: | Huang, R.H, Fatma, S, Chakravarti, A. | | Deposit date: | 2021-08-20 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular mechanisms of the CdnG-Cap5 antiphage defense system employing 3',2'-cGAMP as the second messenger.
Nat Commun, 12, 2021
|
|
7RWS
 
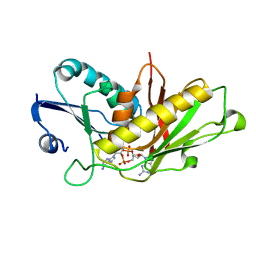 | |
4N14
 
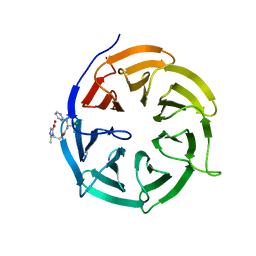 | | Crystal structure of Cdc20 and apcin complex | | Descriptor: | 2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethyl [(1R)-2,2,2-trichloro-1-(pyrimidin-2-ylamino)ethyl]carbamate, Cell division cycle protein 20 homolog | | Authors: | Luo, X, Tian, W, Yu, H. | | Deposit date: | 2013-10-03 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C.
Nature, 514, 2014
|
|
7JY5
 
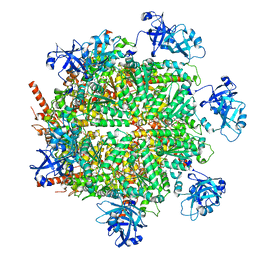 | | Structure of human p97 in complex with ATPgammaS and Npl4/Ufd1 (masked around p97) | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Seesaw conformations of Npl4 in the human p97 complex and the inhibitory mechanism of a disulfiram derivative.
Nat Commun, 12, 2021
|
|
8URN
 
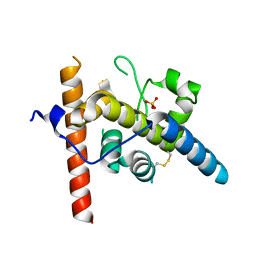 | | Crystal structure of EscI(51-87)-linker-EtgA(18-152) fusion protein | | Descriptor: | EscI inner rod protein type III secretion system,EtgA protein, SULFATE ION | | Authors: | van den Akker, F. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insights into peptidoglycan glycosidase EtgA binding to the inner rod protein EscI of the type III secretion system via a designed EscI-EtgA fusion protein.
Protein Sci., 33, 2024
|
|
4GL2
 
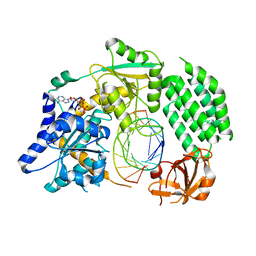 | | Structural Basis for dsRNA duplex backbone recognition by MDA5 | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RNA (5'-R(*AP*UP*CP*CP*GP*CP*GP*GP*CP*CP*CP*U)-3'), ... | | Authors: | Wu, B, Hur, S. | | Deposit date: | 2012-08-13 | | Release date: | 2013-01-09 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (3.557 Å) | | Cite: | Structural Basis for dsRNA Recognition, Filament Formation, and Antiviral Signal Activation by MDA5.
Cell(Cambridge,Mass.), 152, 2013
|
|
8JBR
 
 | | Structure of McyA2-CAPCP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, McyA protein | | Authors: | Peng, Y.J. | | Deposit date: | 2023-05-09 | | Release date: | 2024-01-24 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Modular catalytic activity of nonribosomal peptide synthetases depends on the dynamic interaction between adenylation and condensation domains.
Structure, 32, 2024
|
|
5YKF
 
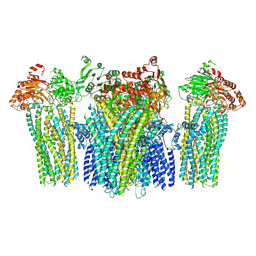 | |
5YKG
 
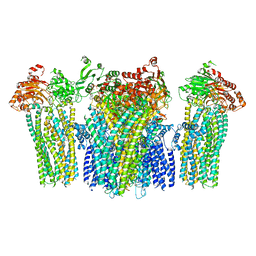 | |
8GF1
 
 | |
8GFC
 
 | |
8GFF
 
 | |
8GFE
 
 | |
8GFJ
 
 | |
8GFG
 
 | | Crystal structure of soluble lytic transglycosylase Cj0843 of Campylobacter jejuni in complex with Z7912 inhibitor | | Descriptor: | (3S,3aR,5S,6S,6aS)-2-oxohexahydro-2H-3,5-methanocyclopenta[b]furan-6-yl 2-acetamido-2-deoxy-beta-D-glucopyranoside, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2023-03-08 | | Release date: | 2023-05-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Exploring the inhibition of the soluble lytic transglycosylase Cj0843c of Campylobacter jejuni via targeting different sites with different scaffolds.
Protein Sci., 32, 2023
|
|
8GFM
 
 | |
8GFH
 
 | |
