7XO8
 
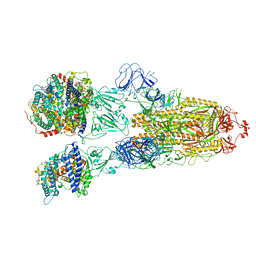 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with three human ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XO6
 
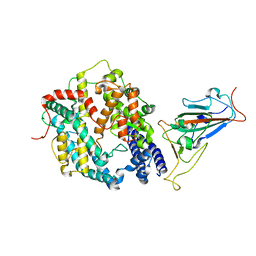 | | SARS-CoV-2 Omicron BA.1 Variant RBD with mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XOC
 
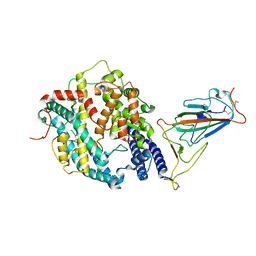 | | SARS-CoV-2 Omicron BA.2 Variant RBD complexed with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XOB
 
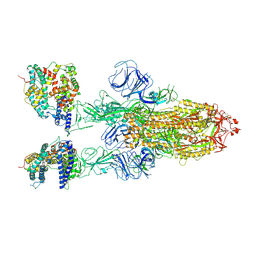 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with two mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
8IWJ
 
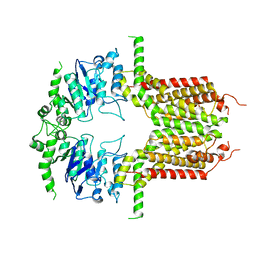 | |
8IWN
 
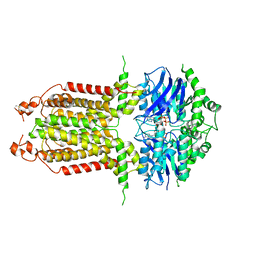 | | ABCG25 EQ mutant in ATP-bound state | | Descriptor: | ABC transporter G family member 25, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Sun, L, Liu, X, Ying, W, Liao, L, Wei, H. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | ABCG25 EQ mutant in ATP-bound state
To Be Published
|
|
8IWK
 
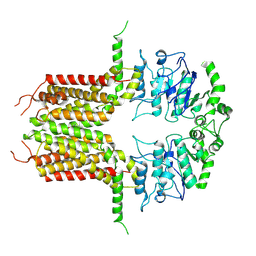 | | ABCG25 Wild Type purified with DDM plus CHS in ABA-bound state | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABC transporter G family member 25 | | Authors: | Sun, L, Liu, X, Ying, W, Liao, L, Wei, H. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | ABCG25 Wild Type purified with DDM plus CHS in ABA-bound state
To Be Published
|
|
8K0Z
 
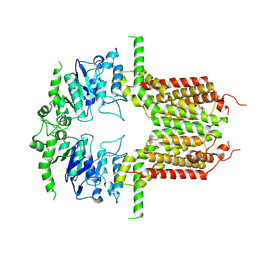 | |
8K0X
 
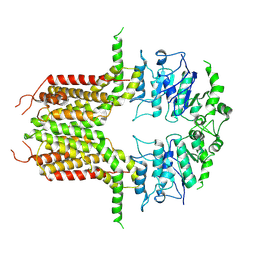 | | ABCG25 Wild Type purified with DDM in the ABA-bound state | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABC transporter G family member 25 | | Authors: | Sun, L, Liu, X, Ying, W, Liao, L, Wei, H. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | ABCG25 Wild Type purified with DDM in the ABA-bound state
To Be Published
|
|
7DFG
 
 | | Structure of COVID-19 RNA-dependent RNA polymerase bound to favipiravir | | Descriptor: | 6-fluoro-3-oxo-4-(5-O-phosphono-beta-D-ribofuranosyl)-3,4-dihydropyrazine-2-carboxamide, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Li, Z, Zhou, Z, Yu, X. | | Deposit date: | 2020-11-08 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for repurpose and design of nucleotide drugs for treating COVID-19
To Be Published
|
|
7DFH
 
 | |
7DOK
 
 | |
7DOI
 
 | |
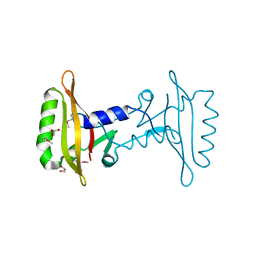
3ICY
 
 | |
3NRT
 
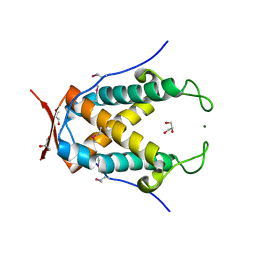 | | The crystal structure of putative ryanodine receptor from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative ryanodine receptor | | Authors: | Wu, R, Zhang, R, James, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | he crystal strucutre of putative ryanodine receptor from Bacteroides thetaiotaomicron VPI-5482
To be Published
|
|
8JBG
 
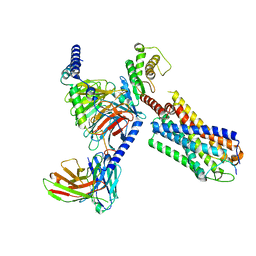 | | Neurokinin B bound to active human neurokinin 3 receptor in complex with Gq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein Gq (G324), ... | | Authors: | Sun, W.J, Yang, F, Zhang, H.H, Yuan, Q.N, Yin, W.C, Shi, P, Eric, X, Tian, C.L. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into neurokinin 3 receptor activation by endogenous and analogue peptide agonists.
Cell Discov, 9, 2023
|
|
8JBH
 
 | | Substance P bound to active human neurokinin 3 receptor in complex with Gq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein Gq subunit alpha (G324), ... | | Authors: | Sun, W.J, Yang, F, Zhang, H.H, Yuan, Q.N, Yin, W.C, Shi, P, Eric, X, Tian, C.L. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into neurokinin 3 receptor activation by endogenous and analogue peptide agonists.
Cell Discov, 9, 2023
|
|
8JBF
 
 | | Senktide bound to active human neurokinin 3 receptor in complex with Gq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein Gq subunit alpha, ... | | Authors: | Sun, W.J, Yang, F, Zhang, H.H, Yuan, Q.N, Yin, W.C, Shi, P, Eric, X, Tian, C.L. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into neurokinin 3 receptor activation by endogenous and analogue peptide agonists.
Cell Discov, 9, 2023
|
|
8IRU
 
 | | Dopamine Receptor D4R-Gi-Rotigotine complex | | Descriptor: | CHOLESTEROL, D(4) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, P, Huang, S, Zhuang, Y, Mao, C, Zhang, Y, Wang, Y, Li, H, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural genomics of the human dopamine receptor system.
Cell Res., 33, 2023
|
|
6H1U
 
 | | GlnH bound to Asp, Mycobacterium tuberculosis | | Descriptor: | ASPARTIC ACID, Probable glutamine-binding lipoprotein GlnH (GLNBP) | | Authors: | O'Hare, H.M, Wallis, R, Lin, L.Y, Newland-Smith, Z. | | Deposit date: | 2018-07-12 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | An Aspartate-Specific Solute-Binding Protein Regulates Protein Kinase G Activity To Control Glutamate Metabolism in Mycobacteria.
MBio, 9, 2018
|
|
6H20
 
 | | GlnH bound to Asn, Mycobacterium tuberculosis | | Descriptor: | ASPARAGINE, Probable glutamine-binding lipoprotein GlnH (GLNBP) | | Authors: | O'Hare, H.M, Wallis, R, Lin, L.Y, Newland-Smith, Z. | | Deposit date: | 2018-07-12 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Aspartate-Specific Solute-Binding Protein Regulates Protein Kinase G Activity To Control Glutamate Metabolism in Mycobacteria.
MBio, 9, 2018
|
|
6H2T
 
 | | GlnH bound to Glu, Mycobacterium tuberculosis | | Descriptor: | GLUTAMIC ACID, Probable glutamine-binding lipoprotein GlnH (GLNBP) | | Authors: | O'Hare, H.M, Wallis, R, Lin, L.Y, Newland-Smith, Z. | | Deposit date: | 2018-07-16 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | An Aspartate-Specific Solute-Binding Protein Regulates Protein Kinase G Activity To Control Glutamate Metabolism in Mycobacteria.
MBio, 9, 2018
|
|
2KYM
 
 | |
4CDX
 
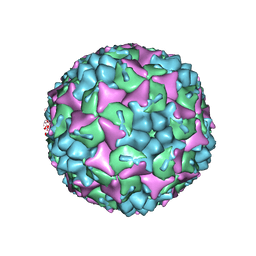 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP12 | | Descriptor: | 1-(5-((3'-METHYL-[1,1'-BIPHENYL]-4-YL)OXY)PENTYL)-3-(, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-07 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDW
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP4 | | Descriptor: | 1-[(3S)-5-(4-iodanylphenoxy)-3-methyl-pentyl]-3-pyridin-4-yl-imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-06 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
