9JJG
 
 | | Cryo-EM structure of RHDV GI.2 virion | | Descriptor: | Genome polyprotein | | Authors: | Ruan, Z, Shao, Q, Song, Y, Hu, B, Fan, Z, Wei, H, Liu, Y, Wang, F, Fang, Q. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Near-atomic structures of RHDV reveal insights into capsid assembly and different conformations between mature virion and VLP.
J.Virol., 2024
|
|
9JJI
 
 | | Local refinement of RHDV GI.2 T=1 VLP | | Descriptor: | Capsid protein | | Authors: | Ruan, Z, Shao, Q, Song, Y, Hu, B, Fan, Z, Wei, H, Liu, Y, Wang, F, Fang, Q. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Near-atomic structures of RHDV reveal insights into capsid assembly and different conformations between mature virion and VLP.
J.Virol., 2024
|
|
9JJJ
 
 | | Cryo-EM structure of a T=3 VLP of RHDV GI.2 | | Descriptor: | Capsid protein | | Authors: | Ruan, Z, Shao, Q, Song, Y, Hu, B, Fan, Z, Wei, H, Liu, Y, Wang, F, Fang, Q. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Near-atomic structures of RHDV reveal insights into capsid assembly and different conformations between mature virion and VLP.
J.Virol., 2024
|
|
9JJH
 
 | | Cryo-EM structure of a T=1 VLP of RHDV GI.2 with N-terminal 1-37 residues truncated | | Descriptor: | Capsid protein | | Authors: | Ruan, Z, Shao, Q, Song, Y, Hu, B, Fan, Z, Wei, H, Liu, Y, Wang, F, Fang, Q. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Near-atomic structures of RHDV reveal insights into capsid assembly and different conformations between mature virion and VLP.
J.Virol., 2024
|
|
4GAE
 
 | | Crystal structure of plasmodium dxr in complex with a pyridine-containing inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, ... | | Authors: | Diao, J, Xue, J, Cai, G, Deng, L, Song, Y. | | Deposit date: | 2012-07-25 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antimalarial and Structural Studies of Pyridine-containing Inhibitors of 1-Deoxyxylulose-5-phosphate Reductoisomerase.
ACS Med Chem Lett, 4, 2013
|
|
4S0J
 
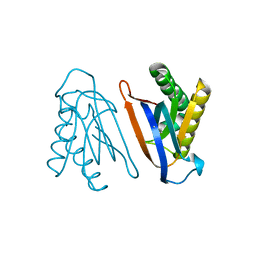 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: 11BIF, 42F, 79S, and 123V mutant | | Descriptor: | Threonine--tRNA ligase | | Authors: | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2014-12-31 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
4S03
 
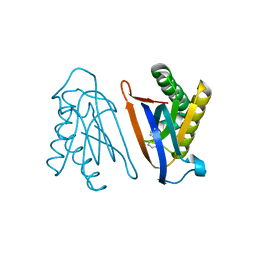 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: I11BIF, Y79I, and F123A mutant | | Descriptor: | Threonine--tRNA ligase | | Authors: | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2014-12-30 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
4S0L
 
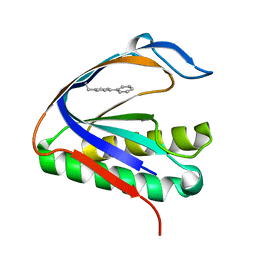 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: I11BIF, Y79V, and F123V mutant | | Descriptor: | Threonine--tRNA ligase | | Authors: | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2014-12-31 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
4S02
 
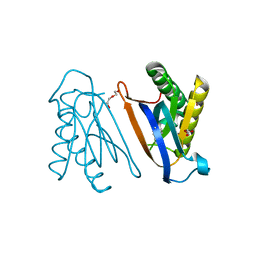 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: I11BIF, F42W, Y79A, and F123Y mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Threonine--tRNA ligase | | Authors: | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2014-12-30 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
4S0K
 
 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: 11BIF, 42F, 79V, and 123A mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Threonine--tRNA ligase | | Authors: | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2014-12-31 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
4S0I
 
 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: 11BIF, 42F, 79S, and 123A mutant | | Descriptor: | Threonine--tRNA ligase | | Authors: | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2014-12-31 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
8EY2
 
 | | Cryo-EM structure of SARS-CoV-2 Main protease C145S in complex with N-terminal peptide | | Descriptor: | 3C-like proteinase | | Authors: | Noske, G.D, Song, Y, Fernandes, R.S, Oliva, G, Godoy, A.S. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An in-solution snapshot of SARS-COV-2 main protease maturation process and inhibition.
Nat Commun, 14, 2023
|
|
4XS3
 
 | | Crystal structure of a metabolic reductase with (E)-1-benzyl-5-((1-methyl-5-oxo-2-thioxoimidazolidin-4-ylidene)methyl)pyridin-2(1H)-one | | Descriptor: | (E)-1-benzyl-5-((1-methyl-5-oxo-2-thioxoimidazolidin-4-ylidene)methyl)pyridin-2(1H)-one, Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, B, Wu, F, Jiang, H, Kogiso, M, Yao, Y, Zhou, C, Li, X, Song, Y. | | Deposit date: | 2015-01-21 | | Release date: | 2016-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.291 Å) | | Cite: | Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds.
J.Med.Chem., 58, 2015
|
|
4XRX
 
 | | Crystal structure of a metabolic reductase with (E)-5-((1-methyl-5-oxo-2-thioxoimidazolidin-4-ylidene)methyl)pyridin-2(1H)-one | | Descriptor: | 5-[(E)-(1-methyl-5-oxo-2-thioxoimidazolidin-4-ylidene)methyl]pyridin-2(1H)-one, Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, B, Wu, F, Jiang, H, Kogiso, M, Yao, Y, Zhou, C, Li, X, Song, Y. | | Deposit date: | 2015-01-21 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds.
J.Med.Chem., 58, 2015
|
|
7ME0
 
 | | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 6.0 | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Song, Y, Nakamura, A.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliva, G. | | Deposit date: | 2021-04-06 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5J2Y
 
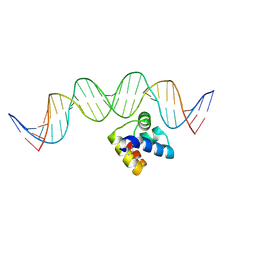 | | Molecular insight into the regulatory mechanism of the quorum-sensing repressor RsaL in Pseudomonas aeruginosa | | Descriptor: | DNA (26-MER), Regulatory protein | | Authors: | Zhao, J, Gan, J, Zhang, J, Kang, H, Kong, W, Zhu, M, Li, F, Song, Y, Qin, J, Liang, H. | | Deposit date: | 2016-03-30 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Pseudomonas aeruginosa RsaL bound to promoter DNA reaffirms its role as a global regulator involved in quorum-sensing.
Nucleic Acids Res., 45, 2017
|
|
8U1J
 
 | |
3RAS
 
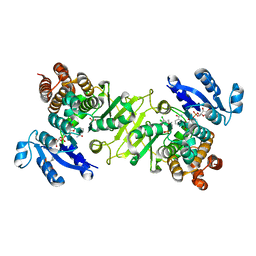 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) complexed with a lipophilic phosphonate inhibitor | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-(N-HYDROXYACETAMIDO)-1-(3,4-DICHLOROPHENYL)PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Diao, J, Deng, L, Prasad, B.V.V, Song, Y. | | Deposit date: | 2011-03-28 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Inhibition of 1-deoxy-D-xylulose-5-phosphate reductoisomerase by lipophilic phosphonates: SAR, QSAR, and crystallographic studies.
J.Med.Chem., 54, 2011
|
|
3SR4
 
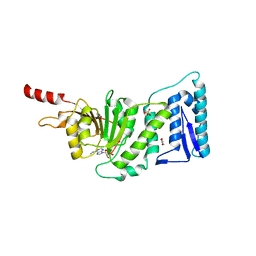 | | Crystal Structure of Human DOT1L in Complex with a Selective Inhibitor | | Descriptor: | (2S)-2-azanyl-4-[[(2S,3S,4R,5R)-5-[6-(methylamino)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Diao, J, Chen, P, Yao, Y, Prasad, B.V.V, Song, Y. | | Deposit date: | 2011-07-06 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective Inhibitors of Histone Methyltransferase DOT1L: Design, Synthesis, and Crystallographic Studies.
J.Am.Chem.Soc., 133, 2011
|
|
8YB4
 
 | | Pfr conformer of Arabidopsis thaliana phytochrome B in complex with phytochrome-interacting factor 6 | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, phytochrome B, phytochrome-interacting factor 6 | | Authors: | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 2024
|
|
9IUZ
 
 | | Constitutively active mutant(Y276H) of Arabidopsis phytochrome B(phyB) in complex with phytochrome-interacting factor 6(PIF6) | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B, Phytochrome-interacting factor 6 | | Authors: | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | Deposit date: | 2024-07-22 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 2024
|
|
4YLY
 
 | | Crystal structure of peptidyl-tRNA hydrolase from a Gram-positive bacterium, staphylococcus aureus at 2.25 angstrom resolution | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Zhang, F, Song, Y, Li, X, Teng, M.K. | | Deposit date: | 2015-03-06 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of Staphylococcus aureus peptidyl-tRNA hydrolase at a 2.25 angstrom resolution.
Acta Biochim.Biophys.Sin., 47, 2015
|
|
6KNM
 
 | | Apelin receptor in complex with single domain antibody | | Descriptor: | Apelin receptor,Rubredoxin,Apelin receptor, Single domain antibody JN241, ZINC ION | | Authors: | Ma, Y.B, Ding, Y, Song, X, Ma, X, Li, X, Zhang, N, Song, Y, Sun, Y, Shen, Y, Zhong, W, Hu, L.A, Ma, Y.L, Zhang, M.Y. | | Deposit date: | 2019-08-06 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-guided discovery of a single-domain antibody agonist against human apelin receptor.
Sci Adv, 6, 2020
|
|
5CMW
 
 | | Crystal structure of yeast Ent5 N-terminal domain-soaked in KI | | Descriptor: | Epsin-5, GLYCEROL, IODIDE ION | | Authors: | Zhang, F, Song, Y, Li, X, Teng, M.K. | | Deposit date: | 2015-07-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insight into the N-terminal domain of the clathrin adaptor Ent5 from Saccharomyces cerevisiae
Biochem.Biophys.Res.Commun., 477, 2016
|
|
5CB5
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, ... | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
