6OSQ
 
 | | RF1 accommodated state bound Release complex 70S at long incubation time point | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-26 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OST
 
 | | RF2 pre-accommodated state bound Release complex 70S at 24ms | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OSK
 
 | | RF1 accommodated 70S complex at 60 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-01 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
7AJR
 
 | | Virtual screening approach leading to the identification of a novel and tractable series of Pseudomonas aeruginosa elastase inhibitors | | Descriptor: | 2-[2-(1,3-benzothiazol-2-ylmethylcarbamoyl)-1,3-dihydroinden-2-yl]ethanoic acid, Keratinase KP2, SULFATE ION, ... | | Authors: | Leiris, S, Davies, D.T, Sprinsky, N, Castandet, J, Behria, L, Bodnarchuk, M.S, Sutton, J.M, Mullins, T.M.G, Jones, M.W, Forrest, A.K, Pallin, T.D, Karunakar, P, Martha, S.K, Parusharamulu, B, Ramula, R, Kotha, V, Pottabathini, N, Pothukanuri, S, Lemonnier, M, Everett, M. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Virtual Screening Approach to Identifying a Novel and Tractable Series of Pseudomonas aeruginosa Elastase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
5ZWS
 
 | |
6H77
 
 | | E1 enzyme for ubiquitin like protein activation in complex with UBL | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Soudah, N, Padala, P, Hassouna, F, Mashahreh, B, Lebedev, A.A, Isupov, M.N, Cohen-Kfir, E, Wiener, R. | | Deposit date: | 2018-07-30 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An N-Terminal Extension to UBA5 Adenylation Domain Boosts UFM1 Activation: Isoform-Specific Differences in Ubiquitin-like Protein Activation.
J.Mol.Biol., 431, 2019
|
|
6H78
 
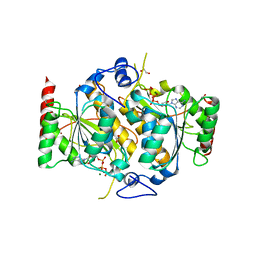 | | E1 enzyme for ubiquitin like protein activation. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Soudah, N, Padala, P, Hassouna, F, Mashahreh, B, Lebedev, A.A, Isupov, M.N, Cohen-Kfir, E, Wiener, R. | | Deposit date: | 2018-07-30 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An N-Terminal Extension to UBA5 Adenylation Domain Boosts UFM1 Activation: Isoform-Specific Differences in Ubiquitin-like Protein Activation.
J.Mol.Biol., 431, 2019
|
|
1PJD
 
 | | Structure and Topology of a Peptide Segment of the 6th Transmembrane Domain of the Saccharomyces cerevisiae alpha-Factor Receptor in Phospholipid Bilayers | | Descriptor: | Pheromone alpha factor receptor | | Authors: | Valentine, K.G, Liu, S.-F, Marassi, F.M, Veglia, G, Nevzorov, A.A, Opella, S.J, Ding, F.-X, Wang, S.-H, Arshava, B, Becker, J.M, Naider, F. | | Deposit date: | 2003-06-02 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure and Topology of a Peptide Segment of the 6th Transmembrane Domain of the Saccharomyces cerevisiae alpha-Factor Receptor in Phospholipid Bilayers
Biopolymers, 59, 2001
|
|
1FYU
 
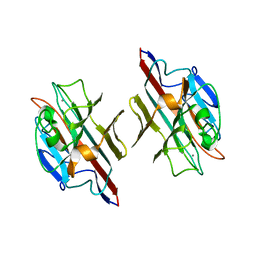 | |
2X6I
 
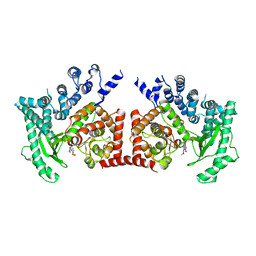 | | THE CRYSTAL STRUCTURE OF THE DROSOPHILA CLASS III PI3-KINASE VPS34 IN COMPLEX WITH PIK-90 | | Descriptor: | N-(2,3-DIHYDRO-7,8-DIMETHOXYIMIDAZO[1,2-C] QUINAZOLIN-5-YL)NICOTINAMIDE, PHOSPHOTIDYLINOSITOL 3 KINASE 59F | | Authors: | Miller, S, Tavshanjian, B, Oleksy, A, Perisic, O, Houseman, B.T, Shokat, K.M, Williams, R.L. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Shaping Development of Autophagy Inhibitors with the Structure of the Lipid Kinase Vps34.
Science, 327, 2010
|
|
2X6H
 
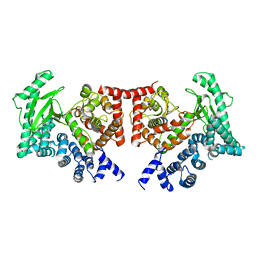 | | THE CRYSTAL STRUCTURE OF THE DROSOPHILA CLASS III PI3-KINASE VPS34 | | Descriptor: | PHOSPHOTIDYLINOSITOL 3 KINASE 59F | | Authors: | Miller, S, Tavshanjian, B, Oleksy, A, Perisic, O, Houseman, B.T, Shokat, K.M, Williams, R.L. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Shaping Development of Autophagy Inhibitors with the Structure of the Lipid Kinase Vps34.
Science, 327, 2010
|
|
2F1F
 
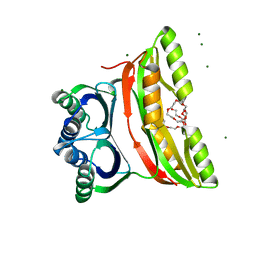 | | Crystal structure of the regulatory subunit of acetohydroxyacid synthase isozyme III from E. coli | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Acetolactate synthase isozyme III small subunit, MAGNESIUM ION, ... | | Authors: | Kaplun, A, Vyazmensky, M, Barak, Z, Chipman, D.M, Shaanan, B. | | Deposit date: | 2005-11-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Regulatory Subunit of Acetohydroxyacid Synthase Isozyme III from Escherichia coli.
J.Mol.Biol., 357, 2006
|
|
2X6F
 
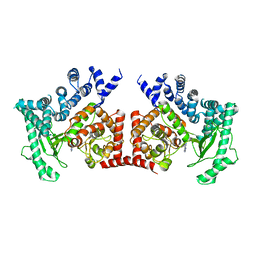 | | THE CRYSTAL STRUCTURE OF THE DROSOPHILA CLASS III PI3-KINASE VPS34 IN COMPLEX WITH 3-METHYLADENINE | | Descriptor: | 6-AMINO-3-METHYLPURINE, PHOSPHOTIDYLINOSITOL 3 KINASE 59F | | Authors: | Miller, S, Tavshanjian, B, Oleksy, A, Perisic, O, Houseman, B.T, Shokat, K.M, Williams, R.L. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Shaping Development of Autophagy Inhibitors with the Structure of the Lipid Kinase Vps34.
Science, 327, 2010
|
|
2X6J
 
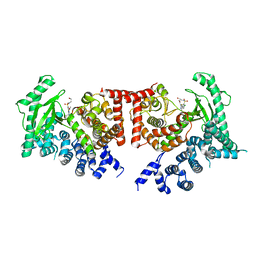 | | THE CRYSTAL STRUCTURE OF THE DROSOPHILA CLASS III PI3-KINASE VPS34 IN COMPLEX WITH PIK-93 | | Descriptor: | N-(5-(4-CHLORO-3-(2-HYDROXY-ETHYLSULFAMOYL)- PHENYLTHIAZOLE-2-YL)-ACETAMIDE, PHOSPHOTIDYLINOSITOL 3 KINASE 59F | | Authors: | Miller, S, Tavshanjian, B, Oleksy, A, Perisic, O, Houseman, B.T, Shokat, K.M, Williams, R.L. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Shaping Development of Autophagy Inhibitors with the Structure of the Lipid Kinase Vps34.
Science, 327, 2010
|
|
2X6K
 
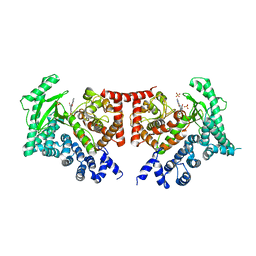 | | THE CRYSTAL STRUCTURE OF THE DROSOPHILA CLASS III PI3-KINASE VPS34 IN COMPLEX WITH PI-103 | | Descriptor: | 3-(4-MORPHOLIN-4-YLPYRIDO[3',2':4,5]FURO[3,2-D]PYRIMIDIN-2-YL)PHENOL, PHOSPHOTIDYLINOSITOL 3 KINASE 59F, SULFATE ION | | Authors: | Miller, S, Tavshanjian, B, Oleksy, A, Perisic, O, Houseman, B.T, Shokat, K.M, Williams, R.L. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Shaping Development of Autophagy Inhibitors with the Structure of the Lipid Kinase Vps34.
Science, 327, 2010
|
|
3HZS
 
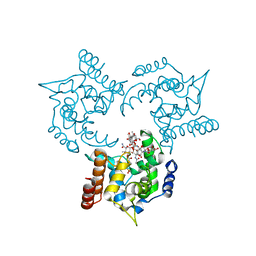 | | S. aureus monofunctional glycosyltransferase (MtgA)in complex with moenomycin | | Descriptor: | MOENOMYCIN, Monofunctional glycosyltransferase, PHOSPHATE ION | | Authors: | Heaslet, H, Miller, A.A, Shaw, B, Mistry, A. | | Deposit date: | 2009-06-24 | | Release date: | 2009-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the active site of S. aureus monofunctional glycosyltransferase (Mtg) by site-directed mutation and structural analysis of the protein complexed with moenomycin
J.Struct.Biol., 167, 2009
|
|
1BCG
 
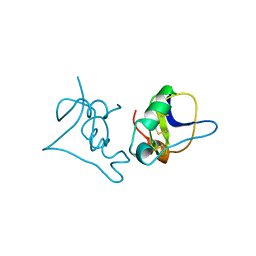 | | SCORPION TOXIN BJXTR-IT | | Descriptor: | TOXIN BJXTR-IT | | Authors: | Oren, D, Froy, O, Amit, E, Kleinberger-Doron, N, Gurevitz, M, Shaanan, B. | | Deposit date: | 1998-04-29 | | Release date: | 1998-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An excitatory scorpion toxin with a distinctive feature: an additional alpha helix at the C terminus and its implications for interaction with insect sodium channels.
Structure, 6, 1998
|
|
3RSW
 
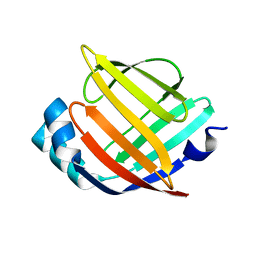 | | Crystal Structure of Heart Fatty Acid Binding Protein (FABP3) | | Descriptor: | Fatty acid-binding protein, heart | | Authors: | Carney, D.F, Shankaran, B, Zwart, P.H, Stebbins, J.W, Prasad, G.S. | | Deposit date: | 2011-05-02 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Crystal Structure of Heart Fatty Acid Binding Protein (FABP3)
To be Published
|
|
2L87
 
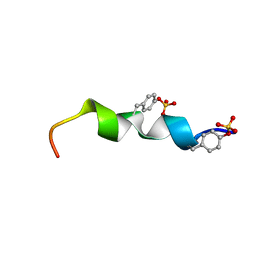 | | The 27-residue N-terminus CCR5-peptide in a ternary complex with HIV-1 gp120 and a CD4-mimic peptide | | Descriptor: | C-C chemokine receptor type 5 | | Authors: | Schnur, E, Noah, E, Ayzenshtat, I, Sargsyan, H, Inui, T, Ding, F.X, Arshava, B, Sagi, Y, Kessler, N, Levy, R, Scherf, T, Naider, F, Anglister, J. | | Deposit date: | 2011-01-06 | | Release date: | 2011-07-27 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The Conformation and Orientation of a 27-Residue CCR5 Peptide in a Ternary Complex with HIV-1 gp120 and a CD4-Mimic Peptide.
J.Mol.Biol., 410, 2011
|
|
5XHN
 
 | | Crystal structure of Frog M-ferritin K104E mutant | | Descriptor: | CHLORIDE ION, Ferritin, middle subunit, ... | | Authors: | Jagdev, M.K, Vasudevan, D. | | Deposit date: | 2017-04-21 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Surface charge dependent separation of modified and hybrid ferritin in native PAGE: Impact of lysine 104
Biochim. Biophys. Acta, 1865, 2017
|
|
5XHM
 
 | | Crystal structure of Frog M-ferritin D40A mutant | | Descriptor: | CHLORIDE ION, Ferritin, middle subunit, ... | | Authors: | Jagdev, M.K, Vasudevan, D. | | Deposit date: | 2017-04-21 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Surface charge dependent separation of modified and hybrid ferritin in native PAGE: Impact of lysine 104
Biochim. Biophys. Acta, 1865, 2017
|
|
5XHI
 
 | | Crystal structure of Frog M-ferritin D38A mutant | | Descriptor: | CHLORIDE ION, Ferritin, middle subunit, ... | | Authors: | Jagdev, M.K, Vasudevan, D. | | Deposit date: | 2017-04-21 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Surface charge dependent separation of modified and hybrid ferritin in native PAGE: Impact of lysine 104
Biochim. Biophys. Acta, 1865, 2017
|
|
5XHO
 
 | | Crystal structure of Frog M-ferritin E135K mutant | | Descriptor: | CHLORIDE ION, Ferritin, middle subunit, ... | | Authors: | Jagdev, M.K, Vasudevan, D. | | Deposit date: | 2017-04-21 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Surface charge dependent separation of modified and hybrid ferritin in native PAGE: Impact of lysine 104
Biochim. Biophys. Acta, 1865, 2017
|
|
5AJK
 
 | |
5AJJ
 
 | |
