5KLT
 
 | | Prototypical P4[M]cNLS | | Descriptor: | Importin subunit alpha-1, Prototypical P4[M]cNLS | | Authors: | Smith, K.M, Forwood, J.K. | | Deposit date: | 2016-06-25 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Contribution of the residue at position 4 within classical nuclear localization signals to modulating interaction with importins and nuclear targeting.
Biochim Biophys Acta Mol Cell Res, 1865, 2018
|
|
1JJA
 
 | |
4RYX
 
 | | Crystal structure of RPE65 in complex with emixustat and palmitate, P6522 crystal form | | Descriptor: | (1R)-3-amino-1-[3-(cyclohexylmethoxy)phenyl]propan-1-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, FE (II) ION, ... | | Authors: | Kiser, P.D, Palczewski, K. | | Deposit date: | 2014-12-17 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular pharmacodynamics of emixustat in protection against retinal degeneration.
J.Clin.Invest., 125, 2015
|
|
4RYY
 
 | |
4RYZ
 
 | |
6GW9
 
 | | Concanavalin A structure determined with data from the EuXFEL, the first MHz free electron laser | | Descriptor: | CALCIUM ION, Concanavalin V, MAGNESIUM ION | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-06-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6H0K
 
 | | Hen egg-white lysozyme structure determined with data from the EuXFEL, the first MHz free electron laser, 7.47 keV photon energy | | Descriptor: | Lysozyme C | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6H0L
 
 | | Hen egg-white lysozyme structure determined with data from the EuXFEL, 9.22 keV photon energy | | Descriptor: | Lysozyme C | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6GWA
 
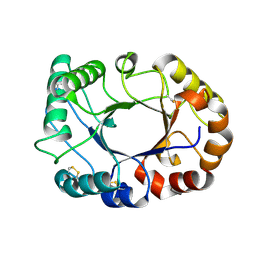 | | Concanavalin B structure determined with data from the EuXFEL, the first MHz free electron laser | | Descriptor: | Concanavalin B | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-06-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6GRZ
 
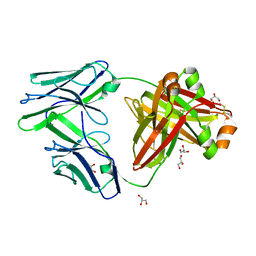 | |
4TNM
 
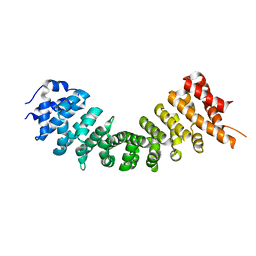 | |
7MPZ
 
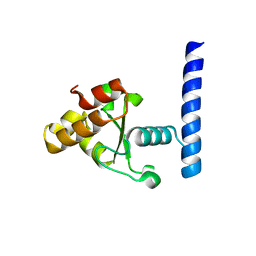 | |
6O56
 
 | | HNH Nuclease from S. pyogenes Cas9 | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1 | | Authors: | Newton, J.C, Lisi, G.P, Jogl, G. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric Motions of the CRISPR-Cas9 HNH Nuclease Probed by NMR and Molecular Dynamics.
J.Am.Chem.Soc., 142, 2020
|
|
8P89
 
 | |
8P88
 
 | |
6RV1
 
 | | human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Cellini, B, Mirco, D. | | Deposit date: | 2019-05-30 | | Release date: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cycloserine enantiomers are reversible inhibitors of human alanine:glyoxylate aminotransferase: implications for Primary Hyperoxaluria type 1.
Biochem.J., 476, 2019
|
|
8F43
 
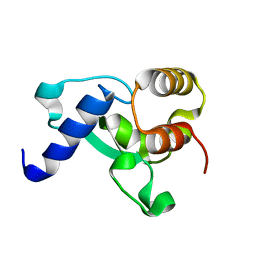 | | HNH Nuclease Domain from G. stearothermophilus Cas9, K597A mutant | | Descriptor: | CRISPR-associated endonuclease Cas9 | | Authors: | D'Ordine, A.M, Belato, H.B, Lisi, G.P, Jogl, G. | | Deposit date: | 2022-11-10 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Disruption of electrostatic contacts in the HNH nuclease from a thermophilic Cas9 rewires allosteric motions and enhances high-temperature DNA cleavage.
J.Chem.Phys., 157, 2022
|
|
8CPE
 
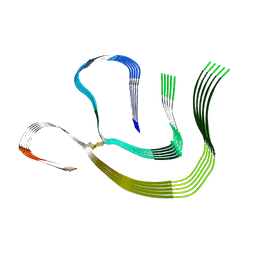 | |
6RV0
 
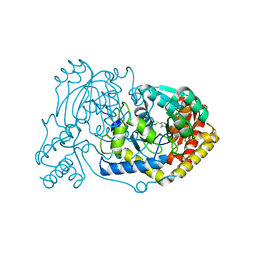 | | human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma); with PMP in the active site | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Cellini, B, Mirco, D. | | Deposit date: | 2019-05-30 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cycloserine enantiomers are reversible inhibitors of human alanine:glyoxylate aminotransferase: implications for Primary Hyperoxaluria type 1.
Biochem.J., 476, 2019
|
|
3BYZ
 
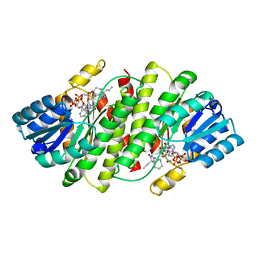 | | 2-Amino-1,3-thiazol-4(5H)-ones as Potent and Selective 11-Hydroxysteroid Dehydrogenase Type 1 Inhibitors | | Descriptor: | (5S)-2-(cyclooctylamino)-5-methyl-5-propyl-1,3-thiazol-4(5H)-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, J, Jordan, S.R, Li, V. | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | 2-amino-1,3-thiazol-4(5H)-ones as potent and selective 11beta-hydroxysteroid dehydrogenase type 1 inhibitors: enzyme-ligand co-crystal structure and demonstration of pharmacodynamic effects in C57Bl/6 mice.
J.Med.Chem., 51, 2008
|
|
6NJ7
 
 | | 11-BETA DEHYDROGENASE ISOZYME 1 IN COMPLEX WITH COLLETOIC ACID | | Descriptor: | (1S,4S,5S,9S)-9-hydroxy-8-methyl-4-(propan-2-yl)spiro[4.5]dec-7-ene-1-carboxylic acid, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Miller, D.J, Rivas, F. | | Deposit date: | 2019-01-02 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanistic Insight on the Mode of Action of Colletoic Acid.
J.Med.Chem., 62, 2019
|
|
6P5G
 
 | | Photoactive Yellow Protein PYP Dark Full | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P4I
 
 | | Photoactive Yellow Protein PYP 10ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-27 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P5F
 
 | | Photoactive Yellow Protein PYP Pure Dark | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P5E
 
 | | Photoactive Yellow Protein PYP 80ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
